CHEMISTRY OF FIREFLY BIOLUMINESCENCE
Bruce R. Branchini
Department of Chemistry
Connecticut College
New London, CT 06320
brbra@conncoll.edu
Introduction
Bioluminescence is an enchanting process in which living organisms convert chemical energy into light. The light results from the oxidation of an organic substrate, a luciferin, catalyzed by an enzyme called a luciferase. In nature, there is an amazing diversity of organisms that emit light including bacteria, fungi, crustaceans, mollusks, fishes and insects (Hastings,1995). While the specific biochemistries of bioluminescence are diverse, all include an enzyme-mediated reaction between molecular oxygen and an organic substrate. It is likely too that all bioluminescence processes involve the formation and breakdown of a four-member ring peroxide or a linear hydroperoxide (Wilson, 1995; Wood,1995). An overview of the chemical and mechanistic aspects of a major bioluminescence process, that of the bioluminescent beetles, will be presented here. The reader is also directed to another review of firefly luciferase (Inouye, 2010).

Figure 1. The North American firefly Photinus pyralis (left), and the Italian firefly Luciola italica (right).
Representing an estimated 2,000 species of luminous beetles (Coeleoptera), are three families: Lampyridae (the true fireflies), Phengodidae (click beetles), and Elateridae (glow-worms). Beginning approximately 60 years ago with the pioneering work of Johns Hopkins University scientists William McElroy, Emil White and Howard Seliger, basic research mainly focused on the common North American firefly Photinus pyralis (Figure 1) has progressed toward a very good understanding of how light is produced by fireflies (DeLuca,1976; McElroy and DeLuca, 1985; White et al., 1971). The recent availability of Luciola cruciata and Photinus pyralis luciferase crystal structures has significantly advanced the understanding of the key structure-function relationships that account for the efficient enzyme-catalyzed emission of light in the firefly (Nakatsu et al., 2006; Sundlov et al., 2012). In turn, the prospects are bright for the continued application of firefly bioluminescence to the already impressive list of medical and pharmaceutical methods (Gould and Subramani, 1998; Contag et al.,1997; Campbell and Sala-Newby, 1993; Kang et al., 2006; Naylor,1999; Gelmini et al., 2000; Kalra et al., 2011; Dragulescu-Andrasi et al.,2011; Branchini et al., 2010), including in vivo imaging to visualize tumors, and for monitoring gene expression and regulation (Figure 2).
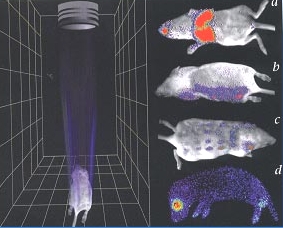
Figure 2. In vivo monitoring of gene expression and infection in mice. The diagram on the left is a representation of the bioluminescence imaging device showing the lens that focuses light from the animal into the CCD detector. On the right, the pseudo color images indicate light intensity (red most and blue least intense). Panel a, bacterial pneumonia is monitored by bioluminescence emission from Salmonella labeled with constitutive expression of the P. luminescens lux operon. Panels b-d show light emission in response to application firefly luciferin concentrations after the introduction of the firefly luciferase gene via a virus (ATP and O2 are endogenous). (from Contag et al., 1998; Nature Medicine 4, 245-247; reproduction permission from Nature Publishing Group).
Biochemical Reactions of Bioluminescence
Firefly bioluminescence is a multi-step process that is outlined in Eqs 1-3 in Figure 3. Luc represents firefly luciferase, ATP is the universal biochemical energy source adenosine triphosphate, PPi is inorganic pyrophosphate, and the structures that correspond to the other abbreviations are shown below. Firefly luciferase has extraordinary specificity for this nucleotide triphosphate. The adenylate is the true substrate of the subsequent oxidative chemistry. In the first step (Eq. 1), luciferase converts firefly D-luciferin (LH2) into the corresponding enzyme-bound luciferyl adenylate. In fact, D-LH2-AMP produced synthetically reacts with oxygen in the presence of luciferase to produce light emission identical to that obtained with the natural substrates D-luciferin and Mg-ATP.
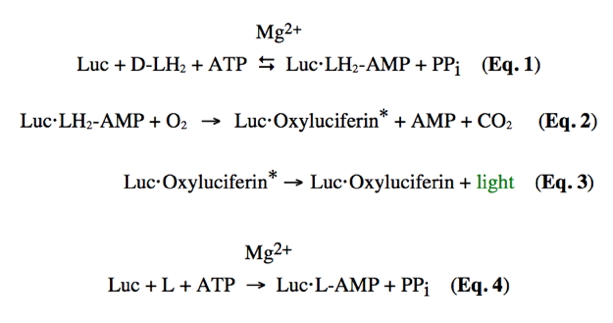
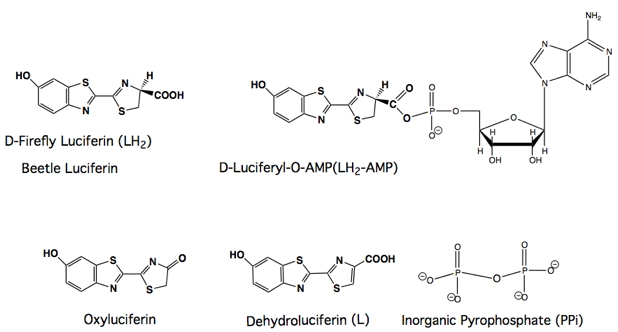
Figure 3. Equations and structures illustrating the reactions catalyzed by firefly luciferase (Luc). The symbol (*) denotes an electronic excited state.
As Equations 2 and 3 indicate, the luciferase enzyme functions as a mono-oxygenase, although it does so in a very unusual manner without the apparent involvement of a metal or cofactor. In some way that has not been yet determined, luciferase amino acid residues are recruited to promote the addition of molecular oxygen to the D-luciferin adenylate, which is then transformed to an electronically excited state oxyluciferin molecule and carbon dioxide, each containing one oxygen atom from molecular oxygen. Visible light emission results from the rapid loss of energy of the excited state oxyluciferin molecule via a fluorescence pathway. The very high quantum yield of ~0.4-0.6 (Niwa et al., 2010a,b) for this process (the emission of a photon from a reacted LH2 molecule) reflects not only efficient catalytic machinery, but also a highly favorable environment that prevents electronically excited state energy loss by non light emitting pathways.
Generally Accepted Mechanism of Bioluminescence
The generally accepted mechanistic details of the overall process of firefly bioluminescence are presented in greater detail below in Figure 4.
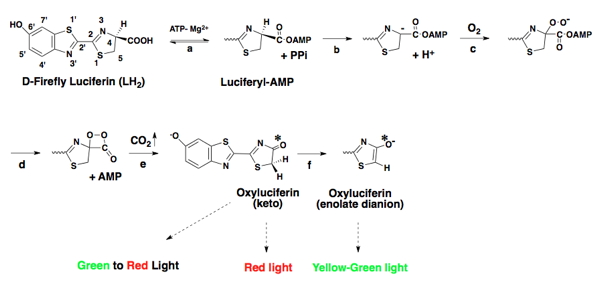
Figure 4. Detailed mechanism of firefly bioluminescence. The symbol (*) denotes an electronic excited state.
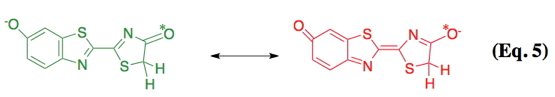
Following the formation of the enzyme-bound luciferyl adenylate (step a), a proton is abstracted from the C-4 carbon of the adenylate by a basic side chain amino acid residue of luciferase (step b). Next, molecular oxygen adds to the newly formed anion (step c); and an electronically excited state oxyluciferin molecule and carbon dioxide are produced (step e) from a highly reactive dioxetanone intermediate (step d). According to the original mechanism based predominantly on chemiluminescence model studies, red light emission (λmax 615 nm), which is observed at pH 6.0, results from the keto form of the emitter. At pH 8, the familiar yellow-green light emission (λmax 560 nm) is produced from the enolate dianion form of the excited state oxyluciferin by a presumed enzymatic assisted tautomerization (step f). However, more recent experimental evidence with a firefly luciferin analog is consistent with the keto form of oxyluciferin alone being capable of producing all of the colors of firefly bioluminescence (Branchini et al., 2002). In nature, beetle luciferases display various colors of light from green (λmax ~535 nm) to red (λmax ~630 nm). Possibly, luciferase modulates emission color by altering the resonance-based charge delocalization of the excited state, as shown below in Eq. 5 (Branchini et al., 2004). There are excellent summaries of the mechanistic proposals on bioluminescence color determination; however, a generally accepted consensus remains elusive (Viviani, 2002; Liu et al., 2008; Hirano et al., 2009; Navizet et al., 2010).
Electronically Excited State Formation
A relatively large amount of excitation energy is required to produce visible light, on the order of 40-70 kcal/mol. The key dioxetanone intermediate shown in the reaction sequence above and in Figure 5 below contains both a strained four-member ring and a weak peroxide bond (O-O). The cleavage of the high energy dioxetanone ring is capable of releasing sufficient energy as a result of the low energy of activation required to cleave the peroxide bond, and the relief of the ring strain inherent in the structure.
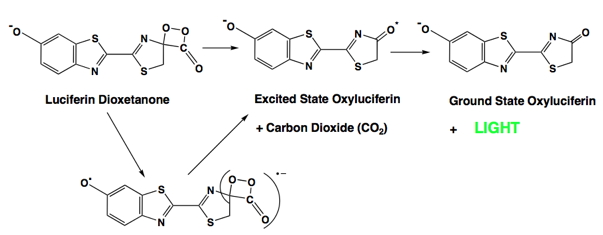
Figure 5. The Chemically Initiated Electron Exchange Luminescence (CIEEL) mechanism for the formation of excited state oxyluciferin in firefly bioluminescence. The key intermediate is shown in brackets.
In the firefly, the energy released is very efficiently directed into the production of an electronically excited state of the bioluminescence product oxyluciferin. Subsequent rapid relaxation of the excited state to the ground state is then accompanied by the emission of a photon of light. One detailed mechanistic view of this process is termed the CIEEL (Chemically Initiated Electron Exchange Luminescence) mechanism (Figure 5) (Schuster et al.,1979; Koo et al.,1978). In firefly bioluminescence, intramolecular electron transfer from the heterocyclic portion of the molecule to the dioxetanone produces a radical ion pair, and the radical anion of carbon dioxide. Next, the back transfer of an electron from the radical anion of carbon dioxide to the radical form of oxyluciferin results in the formation of electronically excited oxyluciferin and carbon dioxide. While the CIEEL mechanism may occur in other bioluminescent systems as well as the firefly (Mager and Tu, 1995), it should be noted that a recent paper presents experimental data that are inconsistent with this mechanism providing sufficient excitation energy for efficient bioluminescence (de Oliveira et al., 2012).
Firefly Luciferase Structure and Mechanistic Functions
The cloning and sequencing of P. pyralis luciferase and similar enzymes from approximately fifteen other beetle species, has revealed that these luciferases are closely related to a large family of non-bioluminescent enzymes that catalyze reactions of ATP with carboxylate substrates to form acyl-adenylates (Conti et al., 1996). The formation of luciferase-bound LH2-AMP and L-AMP (Eqs. 1 and 4) illustrates the chemistry common to this large group of enzymes and it is considered a ligase function. This "acyl-adenylate/thioester-forming" group of enzymes shares an identifying motif, also called a signature sequence (198SSGSTGLPKG207 in Luc), that binds the phosphate groups of ATP contributing to the ligase catalytic function. Recently, Gulick (2009) reviewed the structure-function aspects of the ANL superfamily of adenylating enzymes that includes the acyl-and aryl CoA-synthetases, the adenylation domains of non-ribosomal peptide synthetases (NRPSs). and firefly luciferases. The NRPSs are particularly interesting since they are involved in the biosynthesis of antibiotics by fungi. The acyl CoA-synthetases and NRPSs also generate thioester (e.g., of Coenzyme A) intermediates or products from the initially formed corresponding acyl-adenylates, and these reactions are similar to one suggested to account for the stimulatory effect of Coenzyme A on luciferase activity (Fraga, 2008; Marques and da Silva, 2009). However, CoA is not a required Luc substrate, and the conversion of the adenylate into oxyluciferin and light (Figure 4) is an oxidase function. Therefore, all ANL enzymes catalyze two reactions, and one of them is a ligase reaction. The luciferases differ in that they are the only enzymes in the superfamily that can oxidize adenylates, and it is this reaction that leads to bioluminescence. Thus, luciferase exhibits two distinct enzymatic functions: as a synthetase in the formation of an acyl adenylate, and as a monooxygenase.
The firefly luciferase crystal structure (Figure 6), the first structure of a member of the ANL enzyme family, revealed a unique molecular architecture consisting of a large N-terminal domain (residues 1-436), and a small C-terminal domain (residues 440-550). The structure was solved by Brick and coworkers (Conti et al., 1996) without substrates or products present, so it was not possible to determine which amino acid residues participated in the bioluminescence process. However, based on an analysis of the positions of several strictly conserved residues among a group of enzymes sharing the adenylation function, a general location of the luciferase active site was proposed (Conti et al., 1996).
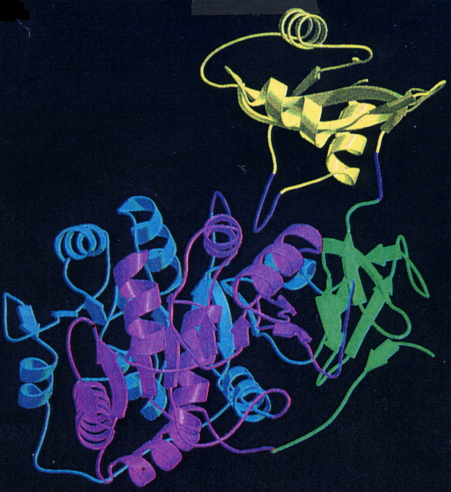
Figure 6. Ribbon diagram of the firefly (P. pyralis) luciferase (Luc) structure. The large N-terminal domain (amino acids 1-436 shown in blue, green and violet) is connected to the smaller C-terminal domain (amino acids 440-550 shown in yellow) through a short hinge peptide (from Conti et al., 1996; reproduction permission from Nature Publishing Group).
Next, the crystal structure of a second member of the adenylate-forming family, the phenylalanine-activating subunit of gramicidin synthetase 1 (PheA) in a complex with phenylalanine, Mg ion and AMP, was reported (Conti et al., 1997). The active site of PheA was determined to be at the interface of the two domains, which were remarkably similar in size and shape to the corresponding domains of luciferase. In the PheA structure, however, the C-terminal domain was rotated 94° and was 5 Å closer to the N-terminal domain than in the luciferase structure. Starting with the two available crystal structures, molecular modeling techniques were used to produce a potential working model of the luciferase active site containing substrates luciferin and Mg-ATP. The model produced in our laboratory is shown below in Figure 7 (Branchini et al., 1998, 2003).
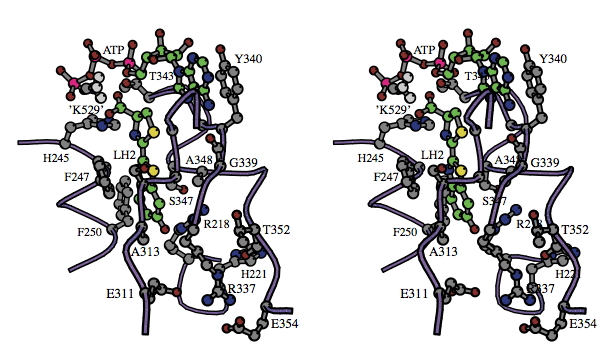
Figure 7. Stereo diagram showing the substrate binding sites suggested by the molecular modeling of luciferase with natural substrates D-LH2 and ATP shown in balls and sticks (carbon atoms are green in both), and Mg2+ ion (not shown). The model was created starting with the luciferase x-ray structure 1LCI, and methyl ammonium ion (labeled 'K529') was used to represent possible interactions of the Lys529 side chain. Traces through the α-carbons of regions V217-H221, H244-T252, H310-L319, and R337-G355 are shown as purple coils. The α-carbons of G246, S314 (and side chain group), G315, G316 and G341 are shown (gray) but are not labeled. The main chain carbonyl groups (oxygen atoms are red) of G339 and T352 also are shown. [The reader is directed to the following site for instructions on how to view stereo diagrams.]
This model proved to be quite useful in the rational design of site-directed mutagenesis-based luciferase structure-function studies, including several related to the determination of bioluminescence color and the characterization of the luciferin-binding site. The idea with this approach is to change amino acids and determine the effect that the change has on the properties of the variant luciferase enzyme containing the change. Systematic mutation of fifteen luciferase amino acid residues and biochemical studies on the resulting mutant proteins has produced data substantiating the view of the firefly luciferin-binding site shown in Figure 7. These and related results can be found in Branchini et al., 2001, 2003; and Leach, 2008.
Based on mutational studies of the luciferases and enzymes in the related acyl adenylate-forming superfamily, a possible mechanism for the luciferase–catalyzed formation of adenylates (Eqs. 1 and 4) is presented in Figure 8.
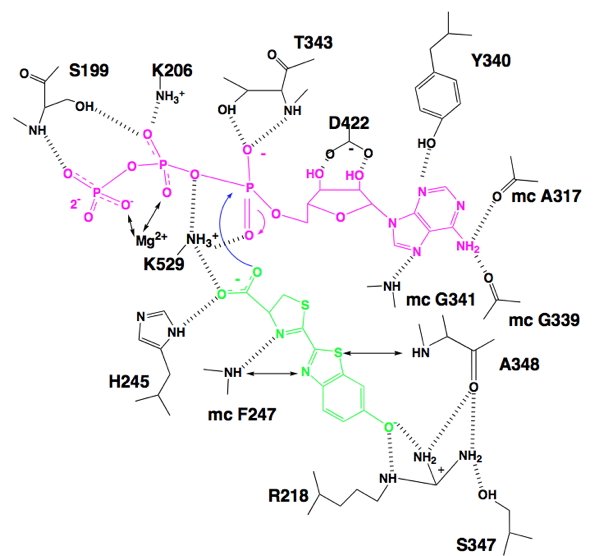
Figure 8. Schematic representation of hydrogen bonding (||||) between Luc and substrates luciferin (green), ATP (violet) and Mg2+ as predicted by molecular modeling. Potential interactions between substrates and atoms in proximity (~2 to ~4 Å) are indicated (↔). For main chain (mc) atoms, only those interacting with substrates are included. The methyl ammonium ion was used to represent possible interactions of the K529 side chain. The curved blue arrows represent the nucleophilic attack of the luciferin carboxylate at the α-phosphorus of ATP, and the corresponding formation of the pentavalent intermediate.
Luciferase residues R218, F247, S347 and A348 are shown making interactions with D-luciferin, fixing its position in the active site. The adenine ring of ATP is held in place by interactions to G339, Y340, G341 and A317, while the side chain carboxylate of D422 is H-bonded to the ribose hydroxyl groups. Residues S199 and K206, highly conserved throughout the acyl adenylate forming superfamily are shown chelating the β- and γ-phosphate portion of ATP. The key residue in the catalytic process mainly responsible for lowering the energy of the transition state is K529, which is probably assisted by T343. The side chain ammonium group of K529 is shown orienting D-luciferin and ATP for a productive bimolecular reaction. The curved arrows show a carboxylate ion oxygen atom of D-luciferin carrying out a nucleophilic attack at the phosphorus atom of the α-phosphate of ATP. A pentavalent transition state is formed that is likely stabilized by electrostatic interactions with the ammonium ion of K529 and H-bonding interactions with the side chain hydroxyl group of T343. Protein stabilization of the transition state accounts for the catalysis of the adenylation reaction. In addition, signature sequence residues S199 and K206 function to remove the inorganic phosphate leaving group as the product adenylate is formed.
The mutagenesis findings, including the importance of the K529 residue in the adenylation half-reaction, were mostly substantiated by the crystallographic results of Kato and coworkers on the structure of firefly luciferase from the Japanese species L. cruciata (Nakatsu et al., 2006) (Figure 9) and a very recent P. pyralis luciferase structure (Sundlov et al., 2012) (Figure 10) in complex with the potent inhibitor DLSA, a non-reactive analog of the luciferyl adenylate. The main differences are that Arg218 does not directly H-bond to the phenol group of DLSA, and Ser347 is H-bonded to the benzothiazole N atom through a bridging water molecule. The wild-type structures represent conformations of luciferase that revealed important interactions for the adenylation reaction.
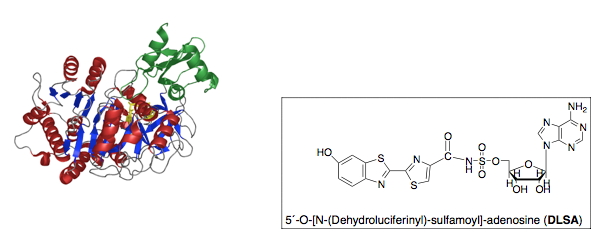
Figure 9. Crystal structure of L. cruciata luciferase in complex with DLSA (Nakatsu et al., 2006). The chemical structure of DLSA is shown.
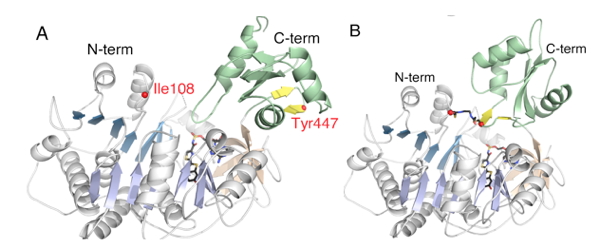
Figure 10. Crystal structures of DLSA complexes of PpyWT in the (A) adenylation and (B) thioester/oxidation conformations, respectively. The structure shown in B was obtained with a P. pyralis variant (Ppy9-) in which residues I108 and Y447 were mutated to Cys and cross-linked with 1,2-bis(maleimido) ethane (BMOE), locking the enzyme in the rotated conformation. The cross-linked luciferase could not produce light with LH2 plus Mg-ATP because it could not make the adenylate; however, bright bioluminescence was observed with synthetic LH2-AMP because it remained capable of catalyzing the oxidative production of excited state oxyluciferin.
Firefly Luciferase Structure and the Domain Alternation Mechanism
According to the domain alternation mechanism (Gulick, 2009), two groups of the ANL superfamily catalyze two partial reactions, adenylation and thioester formation, in two different conformations. This is accomplished by the enzymes forming two active sites through an ~140° rotation of the C-domain around the hinge region connecting the N-domain. Until very recently it was unclear if the luciferases catalyzed the oxidation reaction according to this rotation-requiring mechanism. Mutagenesis evidence did favor this hypothesis as our lab had previously shown that two amino acid residues K443 and K529, which are ~30 Å apart on opposite sides of the C-domain of Luc, are each essential for the catalysis of either the adenylation (K529) or oxidation (K443) half-reactions, but not both (Branchini et al., 2005).
Recently, we constructed a Luc variant called Ppy9- C108/C447 and cross-linked it with BMOE (1,2-bis(maleimido)ethane) trapping the enzyme in a C-domain rotated conformation. The reagent contains reactive groups at each end that formed covalent bonds with the Cys residues that we introduced by mutagenesis on the surface of the N- and C-domains. Since the residues are ~25 Å apart, they could only be cross linked if the C-domain rotated as predicted by the domain alternation mechanism. Importantly, the cross linked Luc variant could not adenylate LH2, but was able to oxidize synthetic LH2-AMP as expected for an enzyme trapped in the rotated conformation poised to catalyze the oxidation of the adenylate. The existence of the two Luc conformations was directly observed in subsequent crystallographic studies of P. pyralis wild type luciferase and Ppy9- C108/C447 DLSA complexes (Figure 10) as predicted by the domain alternation mechanism (Sundlov et al., 2012).
The mutagenesis and crystallographic studies revealed mechanistic details relating the luciferase structure to its function as an oxidase (Eqs. 2 and 3 in Figure 3). In particular, two structural motifs identified in the acyl-adenylate forming superfamily (340YGLTE344 and 442IKYKGYQV449 in luciferase) play significant roles in the catalysis of the oxidative reactions leading to the emission of light in the firefly. The structural results have confirmed the key role of K443 and identified a tunnel that provides molecular O2 access to the active site only in the second partial reaction (Sundlov et al., 2012). Additionally, the results also call into question details of the chemistry of dioxetanone formation (Figure 4), and an alternative hypothesis involving a radical-based mechanism has been proposed (Sundlov et al., 2012). Research continues to be aimed at developing a detailed explanation of just how the firefly oxidizes its substrate to process the familiar lights many associate with beautiful warm summer evenings.
References
Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., and Zimmer, M. (1998) Site-directed mutagenesis of histidine 245 in firefly luciferase: a proposed model of the active site. Biochemistry 37, 15311-15319.
Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., and Portier, N. C. (2001) The role of active site residue arginine 218 in firefly luciferase bioluminescence. Biochemistry 40, 2410-2418.
Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Portier, N. C., Ruggiero, M. C., and Stroh, J. G. (2002) Yellow-green and red firefly bioluminescence from 5,5 dimethyloxyluciferin. J. Am. Chem. Soc. 124, 2112-2113.
Branchini, B. R., Southworth, T. L., Murtiashaw, M. H., Boije, H., and Fleet, S. E. (2003) A mutagenesis study of the putative luciferin binding sirte residues of firefly luciferase. Biochemistry 42, 10429-10436.
Branchini, B. R., Southworth, T. L., Murtiashaw, M. H., Magyar, R. A., Gonzalez, S. A., Ruggiero, M. C., and Stroh, J. G. (2004) An alternative mechanism of bioluminescence color determination in firefly luciferase. Biochemistry 43, 7255- 7262.
Branchini, B. R., Southworth, T. L., Murtiashaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., and Zimmer, M. (2005) Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry 44, 1385-1393.
Branchini, B. R., Ablamsky, D. M., Davis, A. L., Southworth, T. L., Butler, B., Fan, F., Jathoul, A. P., and Pule, M. A. (2010) Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal. Biochem. 396, 290-297.
Branchini, B. R., Rosenberg, J. C., Fontaine, D. M., Southworth, T. L., Behney, C. E., and Uzasci, L. (2011) Bioluminescence is produced from a trapped firefly luciferase conformation predicted by the domain alternation mechanism. J. Am. Chem. Soc. 133, 11088-11091.
Campbell, A. K. and Sala-Newby, G. B. (1993) Bioluminescent and chemiluminescent indicators for molecular signaling and function in living cells. In Fluorescent and Luminescent Probes for Biological Activity (Mason, W. T., Ed.), pp 58-82, Academic Press, London.
Contag, C. H., Spilman, S. D., Contag, P. R., Oshiro, M., Eames, B., Dennery, P., Stevenson, D. K., and Benaron, D. A. (1997) Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol. 66, 523-531.
Conti, E., Franks, N. P., and Brick, P. (1996) Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4, 287- 298.
Conti, E., T. Stachelhaus, Marahiel, M. A., and Brick, P. (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16, 4174-4183.
DeLuca, M. (1976) Firefly luciferase. Adv. Enzymol. 44, 37-68.
Dragulescu-Andrasi, A., Chan, C. T., De, A., Massoud, T. F., and Gambhir, S. S. (2011) Bioluminescence resonance energy transfer (BRET) imaging of protein protein interactions within deep tissues of living subjects. Proc. Natl. Acad. Sci. USA 108, 12060-12065.
Fraga, H. (2008) Firefly Luminescence: a historical perspective and recent developments. Photochem. Photobiol. Sci. 7, 146-158.
Gelmini, S., Pinzani, P., and Pazzagli, M. (2000) Luciferase gene as reporter: comparison with the CAT gene and use in transfection and microinjection of mammalian cells. Methods Enzymol. 305, 557-576.
Gould, S. J. and Subramani, S. (1988) Firefly luciferase as a tool in molecular and cell biology. Anal. Biochem. 175, 5-13.
Gulick, A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811-827.
Hastings, J. W. (1995) Bioluminescence. In Cell Physiology Source Book (Sperelakis, N., Ed.) pp 665-681, Academic Press, New York. Hirano, T., Hasumi, Y., Ohtsuka, K., Maki, S., Niwa, H., Yamaji, M., and Hashizume, D. (2009) Spectroscopic studies of the light-color modulation mechanism of firefly (beetle) bioluminescence. J. Am. Chem. Soc. 131, 2385-2396.
Inouye, S. (2010) Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell. Mol. Life Sci. 67, 387-404.
Kalra, J., Anantha, M., Warburton, C., Waterhouse, D., Yan, H., Yang, Y. J., Strut, D., Osooly, M., Masin, D., and Bally, M. B. (2011) Validating the use of a luciferase labeled breast cancer cell line, MDA435LCC6, as a means to monitor tumor progression and to assess the therapeutic activity of an established anticancer drug, docetaxel (Dt) alone or in combination with the ILK inhibitor, QLT0267. Cancer Biology & Therapy 11, 826-838.
Kang, S. H., Cho, M.-J. and Kole, R. (1998) Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry 37, 6235-6239.
Koo, J.-Y., Schmidt, S. P., and Schuster, G. B. (1978) Bioluminescence of the firefly: key steps in the formation of the electronically excited state for model systems. Proc. Natl. Acad. Sci. U.S.A. 75, 30-33.
Leach, F. R. (2008) A view of the active site of firefly luciferase. Nat. Prod. Commun. 3, 1437-1448.
Liu, Y. J., De Vico, L., and Lindh, R. (2008) Ab initio investigation on the chemical origin of the firefly bioluminescence. J. Photochem. Photobiol. A. 194, 261-267.
Mager, H. I. X. and Tu, S.-C. (1995) Chemical aspects of bioluminescence. Photochem. Photobiol. 62, 607-614.
Marques, S. M. and Esteves da Silva, J. C. G. (2009) Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. Life 61, 6-17.
McElroy, W. D. and DeLuca, M. (1985) Firefly luminescence. In Chemi- and Bioluminescence (Burr, J. G., Ed.) pp 387-399, Marcel Dekker, New York.
Nakatsu, T., Ichiyama, S., Hiratake, J., Saldanha, A., Kobashi, N., Sakata, K., and Kato, H. (2006) Structural basis for the spectral difference in luciferase bioluminescence. Nature 440, 372-376.
Navizet, I., Liu, Y. J., Ferre, N., Xiao, H. Y., Fang, W. H., and Lindh, R. (2010) Color-tuning mechanism of firefly investigated by multi-configurational perturbation method. J. Am. Chem. Soc. 132, 706-712.
Naylor, L. H. (1999) Reporter gene technology: the future looks bright. Biochem. Pharmacol. 58, 749-757.
Niwa, K., Ichino, Y., and Ohmiya, Y. (2010a) Quantum yield measurements of firefly bioluminescence using a commercial luminometer. Chem. Lett. 39, 291-293.
Niwa, K., Ichino, Y., Kumata, S., Nakajima, Y., Hiraishi, Y., Kato, D., Viviani, V. R., and Ohmiya, Y. (2010b) Quantum yields and kinetics of the firefly bioluminescence reaction of beetle luciferases. Photochem. Photobiol. 86, 1046-1049.
de Oliveira, M. A., Bartoloni, F. H., Augusto, F. A., Ciscato, L. F., Bastos, E. L., and Baader, W. J. (2012) Revision of singlet quantum yields in the catalyzed decomposition of cyclic peroxides. J. Org. Chem. 77, 10537-10544.
Schuster, G. B., Dixon, B., Koo, J.-Y., Schmidt, S. P., and Smith, J. P. (1979) Chemical mechanisms of chemi- and bioluminescence. Reactions of high energy content organic compounds. Photochem. Photobiol. 30, 17-26.
Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., and Gulick, A. M. (2012) Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism. Biochemistry 51, 6493-6495.
Viviani, V. R. (2002) The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci. 59, 1833-1850.
White, E. H., Rapaport, E., Seliger, H. H., and Hopkins, T. A. (1971) The chemi and bioluminescence of firefly luciferin: an efficient chemical production of electronically excited states. Bioorg. Chem. 1, 92-122.
Wilson, T. (1995) Comments on the mechanisms of chemi- and bioluminescence. Photochem. Photobiol. 62, 601-606.
Wood, K. V. (1995) The chemical mechanism and evolutionary development of beetle bioluminescence. Photochem. Photobiol. 62, 662-673.
02/18/08
08/02/13