PHOTORECEPTOR MUTANTS for the STUDY of PHOTORECEPTOR INTERACTIONS
Keara A. Franklin1 and James R. Shinkle2
1School of Biological Sciences, University of Bristol, Woodland Road, Bristol, BS8 1UG, UK
kerry.franklin@bristol.ac.uk
2Department of Biology, Trinity University, San Antonio,
Texas 78212-7200, USA
jshinkle@trinity.edu
Introduction
Effective monitoring of the ambient light environment is fundamental to plant growth and development. Through the action of specialized photoreceptors, plants monitor the quantity, quality and photoperiod of incident light, and use this information to modulate all aspects of their development, from seed germination to plant architecture and onset of flowering. Three principle families of photoreceptor have been identified for light perception in higher plant tissues, the red/far-red (R/FR) light-absorbing phytochromes (PHYs), and the UV-A/blue light (B)-absorbing cryptochromes (CRYs) and phototropins (PHOTs) (reviewed in Quail, 2002). Elucidation of the roles of individual photoreceptors in mediating plant development is, however, confounded by their redundant, synergistic and in some cases, mutually antagonistic mechanisms of action.
This module presents accounts of how loss-of-function mutants in the genes encoding the cryptochromes, phototropins and phytochromes have been used to investigate interactions between these photoreceptors. However, it is worth noting in passing that deliberate modifications of photoreceptor structure through site-directed mutagenesis has led to insights into how individual photoreceptors initiate signal transduction events. Two important examples are: 1) the demonstration that the chromophore domain of CRY2 confers light regulation of the autophosphorylation of C-terminal domain (Shatlin et al., 2002); and 2) the creation of a PHYB protein that activates responses in the absence of a light signal (Su and Lagarias, 2007). Hence, the accounts below only address a part of the successful application of mutants to understanding photoreceptor function.
The central benefit of loss of function mutants can be explained as follows: The cloning of individual photoreceptor genes, combined with the relative simplicity of introducing and expressing such genes in model systems such as Arabidopsis and tobacco, led to a variety of studies of photoreceptor action using transgenic plants. Although these studies provided valuable information, the use of transgenic technology to define roles for individual photoreceptors can present problems, particularly when the method involves expressing genes at unnaturally high levels. The majority of these strategies have used the introduction of genes from different species. This introduces complications arising from different primary structures, and differing stabilities of proteins expressed in cells of another species.
When photoreceptor genes are expressed outside normal regulatory control, and photoreceptors accumulate to higher than normal levels, they may adopt some or all of the functions of other family members. In such circumstances, a particular photoreceptor may also interact with signal transduction components used by other photoreceptors, interfering with normal signaling (Whitelam and Harberd ,1994). The isolation and characterization of photoreceptor-deficient mutants in a variety of plant species has therefore revealed a refined picture of individual photoreceptor functions. Redundancy within photoreceptor families, and the overlapping absorption spectra of red and blue light sensing systems has, however, impeded investigation into mechanisms of photoreceptor co-action in monogenic mutants. The creation of mutants, null for multiple combinations of photoreceptor, has consequently provided a more realistic and unique insight into both individual functions and mechanisms of co-action.
Definitions:
PHYA (no italics) refers to the apoprotein (protein without chromophore)
PHYA (italics) refers to the gene
phyA (no italics) refers to the holoprotein (protein + chromophore)
phyA (italics) refers to the mutant
Photoreceptor Families
Plants monitor red and far-red wavelengths using the phytochrome family of photoreceptors. Phytochromes exist as a homodimer of two independently reversible subunits. Each subunit consists of a polypeptide (~124 kDa) covalently attached to a light-absorbing tetrapyrrole chromophore (Lagarias and Rapport, 1980). Interactions between the chromophore and polypeptide components enable phytochrome to assume two spectrally distinct forms: Pr, which absorbs maximally in the red region of the spectrum (666-668 nm) and Pfr, which absorbs maximally in the far-red region (730 nm). Phytochrome is synthesised as Pr, which is biologically inactive for most phytochrome-mediated responses. Activity is acquired upon phototransformation to the Pfr isomer (Kendrick and Kronenberg, 1994).
In the model species Arabidopsis thaliana, five discrete apophytochrome-encoding genes, PHYA-PHYE, have been isolated and sequenced (Mathews and Sharrock, 1997) and Phytochromes A, B and C are conserved among angiosperms (Matthews et al., 1995). Phytochromes A, B, C and E are evolutionarily divergent proteins, sharing only 46-53% sequence identity, while PHYD encodes an apoprotein that shares 80% sequence identity with PHYB (Clack et al., 1994). Molecular phylogenetic analysis supports the occurrence of four major duplication events in the evolution of phytochrome genes (Figure 1). An initial duplication is believed to have separated PHYA (light labile in the far-red light absorbing (Pfr) form) and PHYC (light stable in the Pfr form) from PHYB/D/E (all light stable in the Pfr form). The subsequent separation of PHYA from PHYC and PHYB/D from PHYE resulted in three sub families: A/C, B/D and E (Smith, 2000).
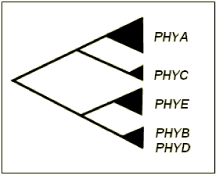
Figure 1. Evolutionary relationship among phytochrome genes in dicotyledonous plants. Phylogenetic analysis has revealed PHYA and PHYC to share a common ancestry, with PHYB, PHYD and PHYE forming a distinct subgroup (Mathews and Sharrock, 1997).
Sensitivity to blue wavelengths is conferred primarily by specialized blue light photoreceptors. In Arabidopsis, these include phototropins (phot 1 and phot 2) and cryptochromes (cry1 and cry2). The former are flavoproteins (proteins covalently attached to flavin chromophore), shown to control directional growth in response to directional light (phototropism), intracellular chloroplast movement in response to photon irradiance, stomatal opening and leaf flattening (Jarillo et al., 2001; Kagawa et al., 2001; Sakai et al., 2001; Sakamoto and Briggs, 2002). Cryptochromes are flavoproteins that show structural similarity to DNA photolyases, and regulate an array of developmental responses throughout plant photomorphogenesis (Briggs and Huala, 1999; Cashmore et al., 1999). A third cryptochrome (cry3) has been identified in Arabidopsis but does not appear to display photoreceptor activity (Kleine et al., 2003).
Seedling Development
The timing of seed germination is regulated, in part, by light signals from the surrounding environment. Induction of germination by red light (R) is mediated primarily by phyB and phyA, whereas induction by far-red light (FR) is mediated solely by phyA (Shinomura et al., 1994, 1996). The retention of R/FR reversible germination responses in phyA phyB double mutants implicated the participation of another phytochrome (Poppe and Schäfer, 1997), a role subsequently assigned to phyE (Hennig et al., 2002).
More recent analyses of mutants deficient in combinations of phyA, phyB, and phyE have shown ambient temperature to modulate the light-regulation of Arabidopsis germination. In this work, phytochrome family members were shown to display altered functional hierarchies at different temperatures (Heschel et al., 2007). At warmer temperatures (>22°C), phyB adopted a prominent role in promoting germination, followed by phyA and phyE. At cooler temperatures (<16°C), phyE displayed functional dominance, with phyB displaying an accessory role (Heschel et al., 2007). The increased functional dominance of phyE over phyB at cooler temperatures parallels observations in flowering inhibition (see 'Flowering'), raising the interesting possibility that phyE abundance may exceed phyB levels in these conditions.
Light signals also act to restrain hypocotyl extension, while stimulating the opening and expansion of cotyledons, and the synthesis of chlorophyll (Figures 2 & 3). The parallel formation of photosynthetic chloroplasts provides plants with the capacity for light harvesting and photoautotrophic growth. These aspects of photomorphogenesis are often collectively referred to as "de-etiolation" processes. Multiple mutant studies in R have revealed all five phytochromes to promote cotyledon development, with phyB performing the predominant role (Franklin et al., 2003a). Under white light (W) conditions, cryptochromes are believed to contribute to this response (Ahmad and Cashmore, 1997).
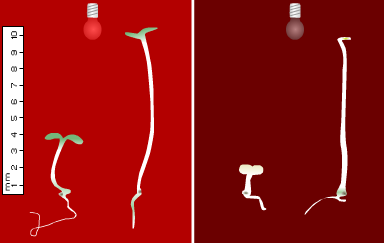
Figure 2. Stem growth and greening during de-etiolation of wild-type and phytochrome mutant Arabidopsis seedlings. For both panels, WT is on the left. Left panel: WT and phyB seedlings grown under continuous Rc for 5 days. Right panel: WT and phyA seedlings grown under continuous FRc for 5 days.
Figure 3 summarizes the behavior of five different phytochrome deficiency genotypes. The isolation of multiple phyC mutants enabled the roles of all five phytochromes to be determined in Arabidopsis seedling de-etiolation (Franklin et al., 2003b; Monte et al., 2003). Mutants deficient in phyC displayed greater hypocotyl elongation than WT controls in R. No additivity was, however, observed in mutants lacking phyB and phyC, suggesting that phyC operates through modulation of phyB function. Despite displaying more sequence similarity to PHYA than PHYB, D or E, no role for phyC in mediating seedling de-etiolation in FR was observed (Franklin et al., 2003b; Monte et al., 2003).
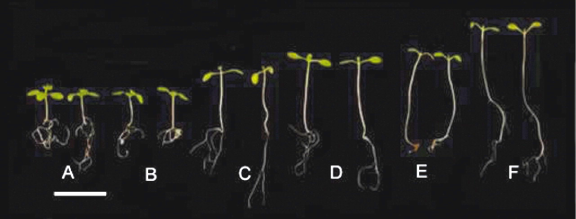
Figure 3. De-etiolation in Arabidopsis seedlings. Genotypes shown are: wild type (A), and mutants in phyA (B), phyB (C), phyBDE (D), phyABE (E) and phyABDE (F). Seedlings were grown in 8 h photoperiods of white light at 120 µmol photons m-2 sec-1. Scale bar represents 5 mm. [Reproduced with permission from Franklin and Quail, 2009]
Mutants deficient in phyB have been characterised in a variety of species including Arabidopsis (Koornneef et al., 1980; Somers et al., 1991), Brassica rapa (Devlin et al., 1992), cucumber (Lopez-Juez et al., 1992), tomato (van Tuinen et al., 1995a), pea (Weller et al., 1995) and Nicotiana plumbagnifolia (Hudson et al., 1997). Analyses of these mutants have revealed a predominant role for phyB in the inhibition of hypocotyl elongation under R (Figure 2, left). In Arabidopsis and tomato, this response is enhanced by synergistic coactions with phyA (Reed et al., 1994; Weller et al., 2000). phyD performs a minor role in hypocotyl inhibition, acting largely in a redundant manner with phyB (Aukerman et al., 1997). The contribution of phyE, however, appears negligible (Devlin et al., 1998).
As noted above, only phyA, phyB and phyC are present in all angiosperms. Grasses such as rice possess just these three phytochromes (Matthews and Sharrock, 1996). While the phytochrome system in rice is simpler than in Arabidopsis, the overall rice genome is more complex. It is only recently that the rice genome has been sequenced (The International Rice Genome Sequencing Project, 2005), and null mutants in the three phytochromes have also only recently been obtained (Takano et al., 2001, 2005). Each class of phy has a role in de-etiolation (Figure 4), and further details of the contributions of the different phys to other aspects of photomorphogenesis are under continuing investigation (Takano et al., 2009).
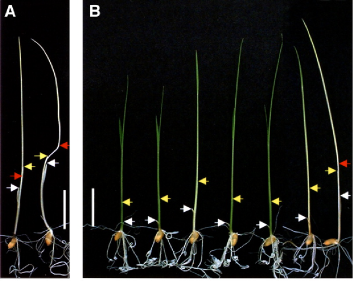
Figure 4. De-etiolation in rice seedlings. Seedlings were grown in darkness (A) or under continuous red light (Rc) (B) for 9 d at 28°C. The fluence rate of Rc was 15 µmol photons m-2 s-1. Two seedlings of Nipponbare (wild type) grown in dark are in (A). In (B) wild-type and single and double phytochrome mutants are aligned for comparison from left to right: wild type, phyA, phyB, phyC, phyA phyC, phyB phyC, and phyA phyB. White and yellow arrows indicate apexes of coleoptiles and first leaves, respectively. Red arrows indicate second nodes. All pictures are the same magnification. Bars = 10 mm. Figure is reproduced with permission from Takano et al. 2005. [DOI: 10.1105/ tpc.105.035889. Copyright The American Society of Plant Biologists.]
The unique role of phyA in mediating hypocotyl inhibition under far-red light (FR) was established through analysis of phyA-deficient mutants in a variety of species including Arabidopsis (Nagatani et al., 1993, Parks and Quail, 1993, Whitelam et al., 1993) (Figure 2, right), tomato (Van Tuinen et al., 1995b) and rice (Takano et al., 2001). In addition, phyA is believed to participate in blue light (B)-sensing mechanisms (Chun et al., 2001; Neff and Chory, 1998; Weller et al., 2001). The experiments implicating phyA in blue light responses used long irradiations, consistent with phyA acting through a High Irradiance Response (see module on Basic Photomorphogenesis).
The majority of photomorphogenic analyses assessing responses in phy mutants have been performed at photon irradiances of <50 µmol m-2 s-1. Growth of seedlings at higher photon irradiances of R (>100 µmol m-2 s-1) has revealed photoprotection of phyA from proteolytic degradation, and considerable phyA activity (Franklin et al., 2007). Under these conditions, phyB mutants displayed markedly greater hypocotyl inhibition and cotyledon expansion than seedlings grown at lower photon irradiances. The irradiance-dependent enhancement of de-etiolation was largely absent in phyA phyB mutants, confirming a significant role for phyA in mediating this response. These findings suggest that, in many natural light environments, where photon irradiances are considerably greater than those achievable in laboratory conditions, the contribution of phyA to seedling establishment may be greater than previously considered, even in light conditions which establish a high proportion of Pfr.
Genes for the B receptors, cry1 and cry2, have been cloned from Arabidopsis and extensively characterized, revealing important regulatory roles throughout plant development (Lin et al., 1996, 1998). A mutant deficient in cry1 has also recently been identified in tomato (Weller et al., 2001). Despite uncertainty over the exact nature of co-action, it is accepted that phyB-mediated de-etiolation involves the interaction between signals from both phytochrome and cryptochrome photoreceptor systems. (Ahmad and Cashmore, 1997; Casal and Mazzela, 1998; Yanovsky et al., 1995). The presence of cry1 was shown to be required for phyB-mediated cotyledon expansion in B and enhanced phyA and phyB-mediated chlorophyll production in R (Neff and Chory, 1998). In a separate study, conditional synergism was observed between cry1 and phyB in hypocotyl inhibition and cotyledon unfolding (Casal and Mazzella, 1998). A functional interaction between cry1 and phyD has also been observed in the W-enhancement of R-mediated hypocotyl inhibition (Hennig et al., 1999). Mutants deficient in phyC have been reported to display hyposensitivity to low photon irradiances of blue light with respect to the regulation of hypocotyl elongation (Franklin et al., 2003b). Under these conditions cry2 function predominates (Lin et al., 1998), suggesting a possible functional interaction between these photoreceptors.
Several kinds of in vitro experiments have demonstrated physical interactions between Arabidopsis CRY1 and PHYA proteins (Ahmad, 1999; Ahmad et al., 1998). A light-dependent direct interaction between cry2 and phyB has also been demonstrated (Más et al., 2000). Direct protein-protein interactions could therefore provide a possible mechanism for photoreceptor co-actions. The R pulse-mediated inhibition of hypocotyl growth in phyB mutants pre-treated with white light has been shown to require the presence of either phyD or cry1. Such responses were reversible by FR, and suggest a functional interaction between these photoreceptors during seedling de-etiolation (Hennig et al., 1999).
Mature Plant Architecture
Light signals regulate plant morphology throughout development. The ability to modulate plant architecture in response to a perceived threat of shading (low R:FR) confers considerable advantage to plants growing in natural communities, and is discussed in the module on Photomorphogenesis in Natural Light Environments. In Arabidopsis, these "shade avoidance" responses are mediated solely by phyB, D and E in a functionally redundant manner (Devlin et al., 1996, 1998, 1999). The analysis of mutants null for multiple photoreceptors has also revealed roles for individual phytochromes in modulating internode elongation and leaf morphology. Mutants deficient in phyB display an elongated phenotype, comparable to that of the shade avoidance syndrome. Such phenotypes suggest a predominant role for phyB in suppressing this response under natural conditions (Whitelam and Devlin, 1997).
Mutants of tomato and Arabidopsis, deficient in phyA, display an almost indistinguishable phenotype from WT plants when grown in white light (van Tuinen et al., 1995b; Whitelam et al., 1993). The role of phyA in modulating mature plant architecture was indicated following an observed reduction in biomass in the phyA phyB double mutant compared with either monogenic mutant (Devlin et al., 1996). Adult Arabidopsis plants structure their leaves in a compact rosette phenotype. The elongated internodes observed in phyA phyB phyE triple mutant plants was the basis on which the phyE mutation was isolated, and led to the proposal that maintenance of the rosette phenotype is regulated, redundantly, by phyA, B and E (Devlin et al., 1998). The elongated appearance of phyA phyB phyD phyE quadruple mutants grown under white light, a phenotype not displayed in phyB phyD phyE triple mutants has supported such a proposal (Franklin et al., 2003a). Figure 5 shows representative plants from these studies.
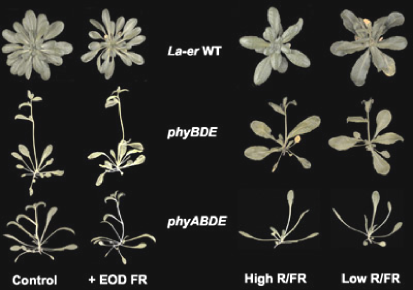
Figure 5. Mutation of phyA affects mature plant architecture. Arabidopsis seedlings grown for 36 days at 16°C under 8h light/16 h dark cycles show two differences between the phyB phyD phyE mutant and the phyA phyB phyD phyE mutant. Under these conditions the loss of phyA causes the loss of the rosette structure, and increases the leaf length to width ratio. [La-er = Landsberg erecta ecotype; EOD = end of day]
Further roles for phyB in modulating plant architecture have recently been proposed in the regulation of stomatal development. Mutants deficient in phyB were shown to display reduced stomatal index (SI) at higher photon irradiances of both W and R (Boccalandro et al., 2009; Casson et al., 2009). Stomatal index represents the ratio of the number of stomata in a given area, divided by the number of stomata and other epidermal cells in the same area, thereby providing an indication of the extent of stomatal differentiation. Increased photon irradiance is well documented to result in increased SI, an adaptation that likely enhances gas exchange during conditions of high photosynthetic activity (Lake et al., 2001). This response displayed significant attenuation in phyB mutants (Casson et al., 2009). A decreased SI was additionally recorded in WT plants subject to End of Day-FR treatment (Boccalandro et al., 2009). The reduced SI observed in phyB mutants was recorded to result in reduced transpiration per unit leaf area, which ultimately enhanced water use efficiency (WUE) in these plants (Boccalandro et al., 2009). The authors propose that phyB-mediated increases in stomatal differentiation serve to enhance photosynthesis in high R:FR/high PAR environments, a strategy that is implemented at the expense of WUE.
Flowering
In many plants, the timing of floral initiation is regulated by the daily duration of light, or photoperiod. Plants in which flowering is accelerated by short days (short-day plants, SDP) generally flower in the autumn to finish reproduction before the adverse temperatures of winter. Plants in which flowering is accelerated by long days (long-day plants, LDP) generally flower in late spring, thus promoting seed set in a more favorable climate. The roles of individual photoreceptors in mediating these responses (termed photoperiodic responses) have been largely inferred from mutant analyses. Two common experimental approaches for studying the regulation of floral initiation are manipulation of daylength using day extension and night break light treatments (Thomas and Vince-Prue, 1997). In day extension experiments, light of low fluence rate is applied at the end of a short-day photoperiod. In night break experiments, a light exposure is given in the middle of a long night, thus mimicking long-day (LD) conditions.
The role of phyA as a promoter of flowering was suggested following observations that a phyA-deficient mutant of Arabidopsis (A LDP) flowered later than wild type (WT) plants in LD (Johnson et al., 1994; Neff and Chory, 1998). Reduced sensitivity of the mutant to night breaks (Reed et al., 1994) and day extensions (Johnson et al., 1994; Neff and Chory, 1998) provided support for such a role. Similar studies in the LDP pea, revealed a phyA-deficient mutant (fun1) to exhibit reduced photoperiodism, and an inability to detect day extensions (Weller et al., 1997).
PhyB is believed to be an inhibitor of flowering. Early flowering and decreased photoperiodic sensitivity were observed in phyB-deficient mutants of Arabidopsis (Goto et al., 1991), pea (Weller and Reid, 1993), and the SDP Sorghum (Childs et al., 1997). It has become apparent, however, than the inhibitory function of phyB in floral initiation is complex. The early flowering phenotype of transgenic Arabidopsis, over-expressing phyB (Bagnall et al., 1995), and the late flowering phenotype of Arabidopsis phyB mutants grown at low temperatures (Halliday et al., 2003), suggest floral initiation to be governed by an interplay of complex regulatory mechanisms. Deficiency of phyD was shown to confer no obvious aberration of floral initiation in Arabidopsis (Aukerman et al., 1997), whereas phyB phyD double mutants flowered earlier than either monogenic parent (Devlin et al., 1999). A similar situation was observed for phyE mutants (Devlin et al., 1998), suggesting inhibition of flowering to be regulated by phyB, phyD and phyE in a functionally redundant manner. Studies of flowering have also provided evidence that a single class of phytochrome can have opposing regulatory roles. When plants are grown under long days, loss of phyA results in later flowering, suggesting a promotory role for this phytochrome (Johnson et al., 1994; Neff and Chory, 1998). However, in the absence of phyD, the additional loss of phyA results in earlier flowering, suggesting an inhibitory role for this phytochrome (Halliday and Whitelam, 2003).
The inductive effect of B on floral initiation in Arabidopsis implied the action of a separate B-absorbing receptor (Mozely and Thomas, 1995). A role for cry2 in the photoperiodic control of flowering was subsequently suggested (Guo et al., 1998). The authors propose that phytochromes mediate the R-dependent inhibition of flowering, whereas cry2 mediates the B-dependent inhibition of phytochrome function. Analysis of the cry2 phyB double mutant has provided support for such a hypothesis (Mockler et al., 1999).
Mutant studies have also revealed roles for phyC in the regulation of Arabidopsis flowering time. An intriguing detail uncovered in these studies is that the contribution of phyC varies among the different ecotypes (geographic varieties traceable to locations where specific genetic stocks were collected) of Arabidopsis. Accelerated flowering was observed in phyC-deficient mutants in the Columbia ecotype when grown in short days (Monte et al., 2003). Comparison of phyA phyC mutants with monogenic parents also revealed a small redundant role for phyC in the detection of long photoperiods with phyA (Monte et al., 2003). No role for phyC in the regulation of flowering was observed in the Wassilewskija ecotype (Franklin et al., 2003b), suggesting the existence of natural variation in phyC function. Quantitative genetic analyses have since suggested that allelic variation at the PHYC locus accounts for considerable latitudinal variation in flowering time, thereby providing support for this notion (Balasubramanian et al., 2006).
As found for germination responses (see 'Seedling Development'), the relative contributions of the different phys to the repression of flowering has been shown to display regulation by ambient growth temperature. When grown at 22°C, phyB mutants are characterized by their early flowering phenotype (Goto et al., 1991; Whitelam and Smith, 1991; Reed et al., 1993). When grown at 16°C, however, similar flowering times were recorded in phyB and WT plants (Halliday et al., 2003). In cooler conditions phyE appears to adopt a dominant role (Halliday and Whitelam, 2003). When grown in short photoperiods at 16°C, phyA phyB phyD mutants flowered with a similar number of rosette leaves to WT plants. Early flowering was, however, observed in phyA phyB phyD phyE mutants, confirming the significance of phyE function in these conditions.
Concluding Remarks
In the Introduction, it was acknowledged that photoreceptor mutants other than nulls have revealed important aspects of photoreceptor function, and in dimensions beyond photoreceptor interaction. As part of a summary perspective it should also be noted that studies using photoreceptor null mutants have likewise provided insight into aspects of photomorphogenesis beyond photoreceptor interaction. As a specific example, cross-talk between light and temperature signaling pathways has been identified in the regulation of Arabidopsis freezing tolerance. In order to survive sub-zero temperatures, many plants require a period of low temperature (<4°C), termed cold acclimation. Exposure to low temperature elevates the expression of a number of genes encoding proteins (collectively referred to as a "regulon") which protect plants against freezing damage through membrane stabilization and the accumulation of compatible solutes (Chinnusumay et al., 2007).
Phytochromes have been shown to perform a role in the regulation of one such regulon, the C-repeat-Binding-Factor (CBF) regulon (Franklin and Whitelam, 2007). Low R:FR treatment at 16°C was shown to elevate expression of both CBF transcription factors and their downstream targets, the COLD-REGULATED (COR) genes, leading to enhanced freezing tolerance. Intriguingly, the linkage of CBFs to downstream target genes was uncoupled at 22°C. Mutant analyses revealed repression of the CBF regulon to be mediated by phyB and phyD in a non-redundant manner (Franklin and Whitelam, 2007). The authors speculate that the cooler temperatures and prolonged twilight reductions in low R:FR experienced by plants growing at Northern latitudes may confer some seasonal protection against sudden freezing snaps during warmer than average autumn months (Linkosalo and Lechowicz, 2006). A more comprehensive review of applications of phytochrome mutants in Arabidopsis to studies of a wide range of aspects of photomorphogenesis was recently published (Franklin and Quail, 2009).
To return to this module's theme, the creation of mutants, null for multiple photoreceptors, has provided valuable insight into functional redundancy and co-actions during plant development. Table 1 shows a summary of roles identified for each class of phytochrome, and which classes of phy mutants were used in obtaining the information. Phylogenetic studies in Arabidopsis have revealed common evolutionary ancestry between phytochromes B,D and E. These form a distinct subgroup and act redundantly to control inhibition of flowering and shade avoidance responses to low R:FR. Redundant interplay between phyB and E has been previously observed in R/FR-reversible seed germination (Hennig et al., 2002), and the maintenance of rosette habit (Devlin et al., 1998), responses with no identifiable role for phyD.
Functional interaction with cry1, however, has been shown to be mediated redundantly by phyB and phyD, with no apparent role for phyE (Hennig et al., 1999). It can therefore be concluded that phyB, D and E are functionally unique photoreceptors, but act redundantly to regulate multiple responses during Arabidopsis development. phyA shares a common ancestry with phyC. A complication has arisen from observations of heterodimerization between phytochromes BCDE that suggests further functional diversification of these photoreceptors (Sharrock and Clack, 2004). A more recent study has suggested obligate heterodimerization of phyC and phyE with other Type II phytochromes (Clack et al., 2009). If this is the case, null mutants in phyB or phyD will also affect functions regulated by phyC and phyE. However, the de-etiolation and survival of phyA phyB phyD phyE mutants through to flowering in R would appear to suggest some activity of phyC, either as homodimers or monomers in vivo (Franklin et al., 2003b, 2007).
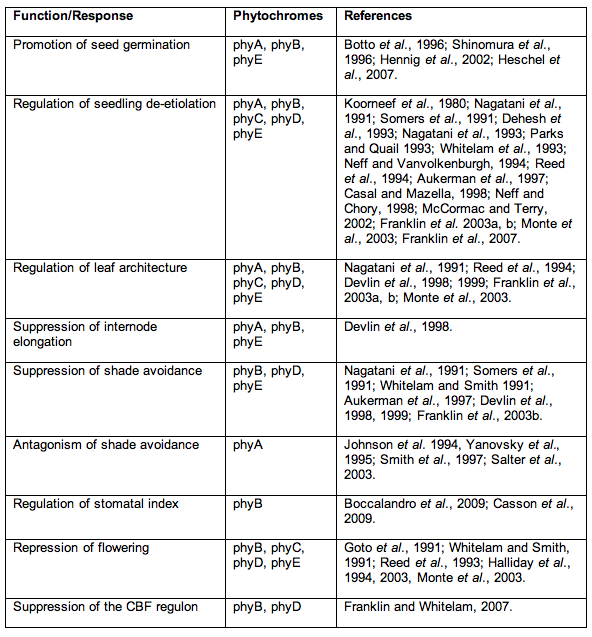
Table 1. Summary of phytochrome functions, elucidated through the analysis of Arabidopsis null mutants. Responses lost in nulls are those that would occur if the lost Phy was in the Pfr form.
In addition to the combined modulation of hypocotyl elongation with other photoreceptors (Reed et al., 1994; Weller et al., 2000), phyA performs regulatory roles in plant architecture (Devlin et al., 1998) and unique functions in FR, conferred by its light lability in the Pfr form. The mechanisms of co-action between phytochrome and blue light-sensing photoreceptors, however, remain to be unequivocally demonstrated. The future isolation of photoreceptor mutants in a variety of species, together with the creation of mutants, deficient in multiple photoreceptors, is therefore a principle objective in elucidating individual photoreceptor functions during plant development.
phyA phyB phyD phyE and phyB phyC phyD phyE plants have been created, containing only phyC and phyA, respectively. The germination, de-etiolation and survival through to flowering of these mutants in both W and R shows the diverse functional capabilities of individual phytochromes; roles which are usually masked by the actions of other family members in WT plants (Figures 3 and 5; Franklin et al., 2003b, 2007). The existence of Arabidopsis quadruple mutants, deficient in four family members, should now enable the implementation of crossing strategies designed to produce a totally phytochrome null plant. The successful construction of a phyA phyB phyC phyD phyE quintuple mutant will, ultimately, address a long unresolved question in photomorphogenesis research, whether phytochromes are an obligate requirement for the germination, growth and reproduction of flowering plants.
Literature Cited
Ahmad M (1999) Seeing the world in red and blue: insight into plant vision and photoreceptors. Curr Opin Plant Biol 2: 230-235.
Ahmad M and Cashmore AR (1997) The blue light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11: 421-427.
Ahmad M, Jarillo J, Smirnova O, and Cashmore AR (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1: 939-948.
Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM and Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317-1326.
Bagnall DJ, King RW, Whitelam GC, Boylan MT, Wagner D and Quail PH (1995) Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol 108: 1495-1503.
Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J and Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nature Genetics 38:711-715.
Briggs WR and Huala E (1999) Blue-light photoreceptors in higher plants. Ann Rev Cell Dev Biol 15: 33-62.
Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ and Casal JJ (2009) Phytochrome B enhances photosynthesis at the expense of water-use-efficiency in Arabidopsis. Plant Physiol 150:1083-1092.
Botto JF, Sanchez RA, Whitelam GC and Casal JJ (1996) Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol 110:439-444.
Casal JJ and Mazzella MA (1998) Conditional synergism between cryptochrome 1 and phytochrome B is shown by analysis of phyA, phyB hy4 simple, double and triple mutants in Arabidopsis. Plant Physiol 118: 19-25.
Cashmore AR, Jarillo JA, Wu YJ and Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760-765.
Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC and Hetherington AM (2009) Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Current Biology 19:229-234.
Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW and Mullet JE (1997) The Sorghum photoperiod sensitivity gene ma3 encodes a phytochrome B. Plant Physiol 113: 611-619.
Chun L, Kawakami A and Christopher DA (2001) Phytochrome A mediates blue light and UV-A dependent chloroplast gene transcription in green leaves. Plant Physiol 125: 1957-1966.
Chinnusumay V, Zhu J, and Zhu JK. (2007) Cold stress regulation of gene expression in plants. Trends in Plant Science 12:444-451.
Clack T, Mathews S and Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes:the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413-427.
Clack T, Shokry A, Moffet M, Liu P, Michael Faul M, and Sharrock RA (2009) Obligate Heterodimerization of Arabidopsis Phytochromes C and E and Interaction with the PIF3 Basic Helix-Loop-Helix Transcription Factor. Plant Cell 21: 786-799.
Dehesh K, Franci C, Parks BM, Seeley KA, Short TW, Tepperman JM and Quail PH (1993) Arabidopsis HY8 locus encodes phytochrome A. Plant Cell 5:1081-1088.
Devlin PF, Halliday KJ, Harberd NP and Whitelam GC (1996) The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: novel phytochromes control internode elongation and flowering time. Plant J 10: 1127-1134.
Devlin PF, Patel SR and Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479-1487.
Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA and Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation and flowering time. Plant Physiol 119: 909-915.
Devlin PF, Rood SB, Somer SE, Quail PH and Whitelam GC (1992) Photophysiology of the elongated internode (ein) mutant of Brassica rapa. The ein mutant lacks a detectable phytochrome B-like polypetide. Plant Physiol 100: 1442-1447.
Franklin KA, Allen T and Whitelam GC (2007) Phytochrome A is an irradiance-dependent red light sensor. Plant J 50:108-117.
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ and Whitelam GC (2003a) Phytochromes B, D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131:1340-1346.
Franklin KA, Davis SJ, Stoddart WM, Vierstra RD and Whitelam GC (2003b) Mutant analyses define multiple roles for phytochrome C in Arabidopsis thaliana photomorphogenesis. Plant Cell 15:1981-1989.
Franklin KA and Whitelam GC (2007) Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nature Genetics 39:1410-1413.
Franklin KA and Quail PH (2009) Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61:11-24.
Goto N, Kumagai T and Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long day plant. Physiol Plant 83:209-215.
Guo H, Yang H, Mockler TC and Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360-1363.
Halliday KJ, Koornneef M and Whitelam GC (1994) Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol 104:1311-1315.
Halliday, KJ and Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol. 131:1913-1920.
Halliday KJ, Salter MG, Thingnaes E and Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33:875-885.
Hennig L, Funk M, Whitelam GC and Schäfer E (1999) Functional interaction of cryptochrome 1 and phytochrome D. Plant J 20: 289-294.
Hennig L, Stoddart WM, Dieterle M, Whitelam GC and Schäfer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128: 194-200.
Heschel MS, Selby J, Butler C, Whitelam GC, Sharrock RA and Donohue K. A new role for phytochromes in temperature-dependent germination. New Phytologist (2007) 174:735-741.
Hudson M, Robson PRH, Kraepiel Y, Caboche M and Smith H (1997) Nicotiana plumbagnifolia hlg mutants have a mutation in a PHYB-type phytochrome gene: they have elongated hypocotyls in red light, but are not elongated in adult plants. Plant J 12: 1091-1101.
Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR and Cashmore AR (2001) Phototropin-related NPL 1 controls chloroplast relocation induced by blue light. Nature 410: 952-954.
Johnson E, Bradley M, Harberd NP and Whitelam GC (1994) Photoresponses of light grown phytochrome A mutants of Arabidopsis. Plant Physiol 105: 141-149.
Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K and Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138-2141.
Kendrick RE and Kronenberg GHM (1994) Photomorphogenesis in plants 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
Kleine T, Lockhart, P and Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35: 93-103.
Koornneef M, Rolff E and Spruitt CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana L. Heyhh. Zeitschrift für pflanzenphysiologie 100: 147-160.
Lagarias JC and Rapport H (1980) Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J Amer Chem Soc 104: 4821-4828.
Lake JA, Quick WP, Beerling DJ and Woodward FI. (2001) Plant development. Signals from mature to new leaves. Nature 411:154.
Lin C, Ahmad M and Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10: 893-902.
Lin C, Yang H, Guo H, Mocker T, Chen J and Cashmore AR (1998) Enhancement of blue light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl Acad Sci USA 95: 7686-7699.
Linkosalo, T. and Lechowicz, M.J. (2006) Twilight far-red treatment advances leaf bud burst of silver birch (Betula pendula) Tree Physiology 26: 1249-1256.
López-Juez E, Nagatani A, Tomizawa K.-I, Deak M, Kern R, Kendrick RE and Furuya M (1992) The cucumber long hypocotyl mutant lacks a light-stable PHYB-like phytochrome. Plant Cell 4: 241-251.
Más P, Devlin P, Panda S and Kay SA (2000) Functional interaction of phytochrome B and crytochrome 2. Nature 408: 207-211.
Mathews S, Lavin M and Sharrock RA (1995) Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Annals of the Missouri Botanic Garden 82:296-321.
Mathews S and Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of loci found in dicot angiosperms. Mol Biol Evol 13: 1141-1150.
Mathews S and Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20: 666-671.
McCormac AC and Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32:549-559.
Mockler TC, Guo H, Yang H, Duong H and Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073-2082.
Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S and Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex cross-talk between phytochrome signaling pathways. Plant Cell 15:1962-1980.
Mozely D and Thomas B (1995) Developmental and photobiological factors affecting photoperiodic induction in Arabidopsis thaliana Heynh. Landsberg erecta. J Ex Bot 46: 173-179.
Nagatani A, Chory J and Furuya M (1991) Phytochrome B is not detectable in the hy3 mutant of Arabidopsis, which is deficient in responding to end-of-day far-red light treatments. Plant and Cell Physiol 32:1119-1112.
Nagatani A, Reed JW and Chory J (1993) Isolation and initial characterisation of Arabidopsis mutants that are deficient in functional phytochrome A. Plant Physiol 102: 269-277.
Neff MM and Van Volkenburgh E (1994) Light-stimulated cotyledon expansion in Arabidopsis seedlings: the role of phytochrome B. Plant Physiol 104:1027-1032.
Neff MM and Chory J (1998) Genetic interaction between phytochrome A, phytochrome B and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27-36.
Parks BM and Quail PH (1993) hy8, a new class of Arabidopsis long hypocotyls mutants deficient in functional phytochrome A. Plant Cell 3: 39-48.
Poppe C and Schäfer E. (1997) Seed germination of Arabidopsis thaliana phyA/phyB double mutants is under phytochrome control. Plant Physiol 114:1487-1492.
Quail PH (2002) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Plant Biol 14: 180-188.
Reed JW, Nagpal P, Poole DS, Furuya M and Chory J (1993) Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147-157.
Reed JW, Nagatani A, Elich T, Fagan M and Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139-1149.
Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M and Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969-6974.
Sakamoto K and Briggs WR (2002) Cellular and subcellular localization of Phototropin 1. Plant Cell 14: 1723-1735.
Salter MG, Franklin KA and, Whitelam GC (2003) Gating of the rapid shade avoidance response by the circadian clock in plants. Nature 426:680-683.
Sharrock, R.A. and Clack, T. (2004). Heterodimerization of type II phytochromes in Arabidopsis. Proc Natl Acad Sci USA 101: 11500-11505.
Shalitin D, Yang HY, Mockler TC, Maymon M, Guo HW, Whitelam GC and Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417:763-67.
Shinomura T, Nagatani A, Chory J and Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104: 363-371.
Shinomura T, Nagatani A, Manzawa H, KubotaM, Watanabe M and Furuya M (1996) Action spectra for phytochrome A and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 8129-8133.
Somers DE, Sharrock RA, Tepperman, JM and Quail PH (1991) The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263-1274.
Smith H, Xu Y and Quail PH (1997) Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiol 114:637-641.
Smith H (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585-591.
Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19:2124-2139.
Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H and Furuya M (2001) Isolation and characterization of rice phytochrome A mutants. Plant Cell 13: 521-534.
Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, and Shinomura T(2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311-3325.
Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, Tanabata T, and Shinomura T (2009) Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice Proc Natl Acad Sci USA 106: 14705-14710.
The International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793-800. Thomas B and Vince-Prue D (1997) Photoperiodism in Plants. Academic Press. London.
Van Tuinen A, Hanhart CJ, Kerckhoffs LHJ, Nagatani A, Boylan MT, Quail PH, Kendrick RE and Koornneef M (1996) Analysis of phytochrome-deficient yellow-green-2 and aurea mutants of tomato. Plant J 9: 173-182.
Van Tuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE and Koorneef M (1995a) A temporarily red light-insensitive mutant of tomato lacks a light-stable B-like phytochrome. Plant Physiol 108: 939-947.
Van Tuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE and Koorneef M (1995b) Far-red light insensitive, phytochrome A-deficient mutants of tomato. Mol. Gen. Genet. 246, 133-141.
Weller JL, Nagatani A, Kendrick RE, Murfet IC and Reid JC (1995) New lv mutants of pea are deficient in phytochrome B. Plant Physiol 108: 525-532.
Weller JL, Murfet IC and JB Reid (1997a) Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 114, 1225-1236.
Weller JL, Perrotta G, Schreuder MEL, van Tuinen A, Koornneef M, Giuliano G and Kendrick RE (2001) Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J 25: 427-440.
Weller JL and Reid J (1993) Photoperiodism and photocontrol of stem elongation in two photomorphogenic mutants of Pisum sativum L. Planta 189: 15-23.
Weller JL, Schreuder MEL, Smith H, Koornneef M and Kendrick RE (2000) Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant J 24: 345-356.
Weller JL, Perrotta G, Schreuder MEL, van Tuinen A, Koornneef M, Giuliano G and Kendrick RE (2001) Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J 25: 427-440.
Whitelam GC and Devlin PF (1997) Roles for different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20: 752-758.
Whitelam GC and Harberd NP (1994) Action and function of phytochrome family members revealed through the study of mutant and transgenic plants. Plant Cell Environ 17: 615-625.
Whitelam GC and Smith H (1991) Retention of phytochrome-mediated shade avoidance responses in phytochrome-deficient mutants of Arabidopsis, cucumber and tomato. J Plant Physiol 39:119-125.
Whitelam GC, Johnson E, Peng J, Carol P, Anderson MC, Cowl JS and Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757-768.
Yanovsky MJ, Casal JJ and Whitelam GC (1995) Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ 18: 788-794.
02/15/10