PHYTOCHROME PHOTORECEPTORS IN CYANOBACTERIA
THE IMPACT OF LIGHT QUALITY ON PHYCOBILISOME BIOSYNTHESIS
Arthur R. Grossman and Devaki Bhaya
The Carnegie Institution for Science
Department of Plant Biology
260 Panama Street, Stanford, CA 94305
arthurg@stanford.edu
dbhaya@stanford.edu
I. INTRODUCTION
Light quality and intensity are critical signals in the environment that allow cells to sense cues that report the time of day, potential intracellular energy status, and help the cell tune its metabolic activities to growth potential. These light signals are sensed as different intensities and qualities (wavelengths) of irradiation. There are numerous photoreceptors that sense light signals, including phytochromes, cryptochromes, phototropins, carotenoids, sensory rhodopsins, as well as the output of the photosynthetic apparatus (redox pressure generated as a consequence of photosynthetic electron transport). Some pigments can also serve to screen out ultraviolet (UV) irradiation or excess excitation energy, or absorb and dissipate the excess energy as heat. In this article, we will focus on the responses of cyanobacteria to the wavelength of light in their environment.
Cyanobacteria are the most ancient oxygenic photosynthetic organisms; inhabiting the Earth for ~3.0 billion years (Schopf, 2002). They are ubiquitous on the planet, flourishing in many extreme environments, such as the oligotrophic oceans, embedded in rocks on the sides of cliffs, and in microbial mats that blanket hotsprings or hypersaline lakes. The cyanobacteria that have been studied in laboratories around the world include mesophiles such Synechococcus PCC7942 and Synechocystis PCC6803, thermophiles such as Thermosynechococcus elongatus, marine organisms such as Prochlorococcus sp. and Synechococcus sp., and some filamentous strains including Anabaena PCC7120, Nostoc punctiforme ATCC 29133 and Fremyella diplosiphon.
One area that has interested us for many years involves the phytochrome-like photoreceptors in cyanobacteria, and more specifically, the role that they and associated regulatory elements play in tailoring the biosynthesis of the major cyanobacterial light harvesting complex, which is called the phycobilisome (PBS), to prevalent wavelengths of light in the environment. This process of molding the characteristics of the PBS to light quality has been designated ‘chromatic adaptation’ (CA), and has been summarized in some recent review articles (Gutu and Kehoe, 2012; Montgomery, 2007).
The type of CA (see Section III.B), and the extent to which an organism acclimates to light quality depends on the genetic characteristics of the organism, which in turn is associated with the environment in which the organism has evolved. However, there are a number of other features of the environment that impact the biogenesis of the photosynthetic apparatus, which includes the light harvesting complexes. For example, levels of PBS in cells can be markedly impacted by the nutrient status of the environment (Collier and Grossman, 1992, 1994; Fujita et al., 1994). It has been known for many years that in addition to the specialized nitrogen storage compound in cyanobacteria called cyanophycin (which is a polymer of aspartate and arginine that is not synthesized on ribosomes), the PBS can serve as a repository for nitrogen that is rapidly accessed when cyanobacterial cells are deprived of nitrogen; cells scavenge the nitrogen in the amino acids that make up the polypeptides of the PBS (see Section III.A for a structural description of the complex).
Indeed, under both nitrogen and sulfur deprivation conditions, the PBS can be rapidly degraded in a highly ordered process that proceeds from the periphery of the rods that make up the PBS, to the core of the complex, which attaches it to the photosynthetic membranes (Grossman et al., 1993a, b; Richaud et al., 2001). Furthermore, regulatory processes appear to integrate the impact of light conditions and nutrient availability on photosynthetic activity, possibly as a consequence of changes in the redox status of the cells (Durnford and Falkowski, 1997; Eberhard et al., 2008; Escoubas et al., 1995; Fujita et al., 1994; van Waasbergen et al., 2002). This physiological flexibility facilitates survival of the organism under rapidly changing environmental conditions.
II. PHYTOCHROME-LIKE PHOTORECEPTORS IN CYANOBACTERIA
Plants contain a number of different phytochromes, which are photoreceptors in which a specific linear tetrapyrrole chromophore, designated phytochromobilin, is covalently linked via the C3 side chain of the A ring to a conserved cysteine residue on the phytochrome apoprotein. The phytochromes of plants have been demonstrated to control numerous physiological processes, including hypocotyl elongation (Nagatani et al., 1991), the shade avoidance response (Casal, 2012; Franklin, 2008), chlorophyll synthesis and the biogenesis of chloroplasts (Frankhauser and Casal, 2004; McCormac and Terry, 2002; Monte et al., 2004), the time of flowering (Osugi et al., 2011), as well as providing signals that integrate with the circadian clock (Millar, 2004; Sellaro et al., 2012; Wenden et al., 2011). Phytochromes of plants have a core sequence with conserved PAS (Per-Arnt-Sim), GAF (cGMP-specific phosphodiesterases, Adenylyl cyclases and FhlA) and PHY (phytochrome) domains and typically exhibit red (R)/far red (FR) photoreversibility.
Phytochromes may be autophosphorylated, and their impact on cell development and physiology may be a consequence of modulating phosphorylation of various regulatory proteins. The GAF domain contains the conserved cysteine residue that bonds with the chromophore. Surprisingly, the phytochrome polypeptide chain forms a true knot with an approximate trefoil (three-lobed) symmetry that creates a novel interface between the PAS and GAF domains, possibly creating a more rigid structure that positions the chromophore binding cysteine as well as chromophore-interacting residues (Wagner et al., 2005); the increased rigidity might reduce the level of de-excitation through protein vibration and domain motion, although advantages of this configuration have not been experimentally demonstrated, and there are many functional phytochrome-like photoreceptors that do not form this knotted structure.
For many years it was controversial whether or not plant phytochromes functioned through protein phosphorylation. This was resolved when Lagarias and colleagues demonstrated that purified plant phytochrome generated in a heterologous system could undergo autophosphorylation through a serine/threonine kinase activity (Yeh and Lagarias, 1998). The state of the art with respect to the structure, function and regulation of plant phytochromes has been summarized in many excellent recent reviews (Auldridge and Forest, 2011; Chen and Chory, 2011; Franklin and Quail, 2010; Hughes, 2010; Kami et al., 2010; Nagatani, 2010; Rockwell and Lagarias, 2010; Rosler et al., 2010; Ulijasz and Vierstra, 2011).
When the entire sequence of the Synechocystis sp. Strain PCC 6803 genome was generated (Kaneko et al., 1996a, b), it became apparent that this cyanobacterium had a number of genes encoding phytochrome-related polypeptides. Over the last 20 years, a significant amount of work has shown that cyanobacteria, other eubacteria and fungi have several chromophore-binding proteins with sequence similarity to plant phytochromes (Rockwell and Lagarias, 2006; Rockwell et al., 2006). While the precise physiological roles of many of these bacterial phytochrome-related proteins are not known, they appear to control growth under certain light conditions (Wilde et al., 1997), carotenoid synthesis and photoprotection (Davis et al., 1999), phototactic movement (Bhaya et al., 2001; Ishizuka et al., 2006; Narikawa et al., 2011; Song et al., 2011; Wilde et al., 2002; Yoshihara et al., 2004; Yoshihara et al., 2000) and chromatic adaptation (CA) (Kehoe and Grossman, 1996); the last of these functions will be discussed in some detail below. Defining the features of phytochrome-like photoreceptors in microbes and the signal transduction processes that they elicit is providing us with new insights into mechanisms involved in light-regulated gene expression, and the evolution of phytochrome structure and function.
Phytochrome-like proteins in bacteria can be grouped into three different subfamilies. One of the subfamilies, represented by Cph1 of Synechocystis PCC6803, has members with features that are similar to that of the plant phytochromes. These photoreceptors have all three domains, PAS, PHY and GAF, which are responsible for the knotted structure (Wagner et al., 2005) that is characteristic of plant phytochrome, although the chromophore to which Cph1 attaches is phycocyanobilin rather than phytochromobilin. Expression of Cph1 has been performed in vitro where it was shown to bind a chromophore, experience a photochromic shift in absorbance, and be capable of protein phosphorylation (Yeh and Lagarias, 1998; Yeh et al., 1997). The photocycle of plant phytochromes and Cph1 show similarities with respect to protonation of the four nitrogens of both the Pr and Pfr configurations of the chromophore (Rohmer et al., 2008; Rohmer et al., 2006; Strauss et al., 2005), as well as a similar photoisomerization of the chromophore; RL absorption by Pr causes a Z to E double bond isomerization at the methine bridge of carbons 15/16, which are positioned between rings C and D of the chromophore.
A representative of the second cyanobacteria subfamily is Cph2 of Synechocystis PCC6803 (Montgomery and Lagarias, 2002; Wilde et al., 2002), although the structure of this protein is fairly complex. Characteristic of other proteins in this group, it does not have the N-terminal PAS domain that is found in both plant phytochromes and Cph1. The absence of this PAS domain eliminates the typical N-terminal, phytochrome ‘knotted structure’. The Cph2 protein also lacks a putative histidine kinase domain that is present in both the C-terminal module of Cph1 and a number of other phytochrome-like proteins. However, Cph2 of Synechocystis PCC6803 exhibits R/FR photoreversibility (Wu and Lagarias, 2000), and its mode of isomerization at carbon 15/16 of the chromophore appears to be essentially identical to that of Cph1. The R/FR photocycle of this photoreceptor is associated with the GAF domain at the N terminus of the protein. Cph2 also exhibits some features that make it distinct from Cph1. The Pfr form of Cph2 decays more rapidly in the dark than that of Cph1 (Anders et al., 2011). Furthermore, Cph2 shows significant variations in its wavelength sensitivities and has been implicated in blue light (BL) photoperception (Fiedler et al., 2005; Wilde et al., 2002).
Interestingly, Cph2 contains two additional GAF domains. One of these might functionally substitute for the PHY domain of Cph1, and the other, located in the C-terminal region of the protein, has homology to tetrapyrrole-binding GAF domains of cyanobacteriochromes (see below). This C-terminal GAF domain appears to bind a phycobilin chromophore that undergoes photoconversion between BL and green light (GL) absorbing states, which can cause it to be tethered to two cysteine residues (Savakis et al., 2012). Furthermore, the Cph2 protein has an EAL and two GGDEF domains (the C-terminal GAF domain is followed by one of the GGDEF domains). GGDEF domains often function as bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP) cyclases, which could cause elevated synthesis of cellular c-di-GMP, while the EAL domain can act as a phosphodiesterases that could cause a reduction in cellular c-di-GMP levels. Hence, photoperception through the chromophores of Cph2 could impact the level of c-di-GMP within the cell, which in turn could impact both motility and biofilm formation (Romling et al., 2005).
The Cph2 protein is a fascinating example of a protein mosaic that has been assembled over evolutionary time from numerous identifiable protein domains that have both sensing and catalytic functions. Integration of multiple functions in this protein may markedly impact the concentration of a small regulatory molecule (c-di-GMP) in the cell, which can then potentially alter a variety of biological processes (including phototaxis). Other examples of cyanobacterial photosensors that lack the PAS domain have been identified in thermophilic Synechococcus sp., and have also been called ‘PAS-less’ Cphs (Ulijasz et al., 2008; Vierstra and Zhang, 2011; Ulijasz et al., 2009).
The third subfamily of cyanobacterial phytochromes is called cyanobacteriochromes or CBCRs. These proteins have a chromophore binding GAF domain (the C-terminal GAF domain in Cph2 is representative of a CBCR chromophore binding site), but no PHY or PAS domain. Furthermore, the absorbance of these photoreceptors may be shifted to the green-violet range of the visible spectrum (Ikeuchi and Ishizuka, 2008; Montgomery and Lagarias, 2002). The first CBCR photoreceptor described, which was identified in F. diplosiphon, was designated RcaE (Regulator of chromatic adaptation E); this protein plays a critical role in chromatic adaptation (Gutu and Kehoe, 2012; Kehoe and Grossman, 1996; Terauchi et al., 2004). Other CBCRs, such as TaxD1, are involved in controlling type IV pilus-dependent phototaxis (Bhaya et al., 2001; Ulijasz et al., 2009), as shown in Figure 1.
Another CBCR that has been studied to some extent includes TePixJ from Thermosynechococcus elongatus (Rockwell et al., 2008; Yoshihara et al., 2004), which exhibits a BL/GL photocycle (Ishizuka et al., 2006; Yoshihara et al., 2004; Yoshihara et al., 2006). These CBCRs are characterized by a second cysteine residue within a conserved sequence motif that is critical for BL/GL sensor function. Ulijasz et al. (2009) have also referred to some of these photosensors in cyanobacteria as 'cyanochromes' and define them as photoreceptors that respond predominantly to BL and GL, and that lack a PHY domain. Some of the CBCR photoreceptors may function as one way photosensors, with the photoproduct exhibiting very slow dark reversion. A number of other CBCRs have been detected by protein sequence alignments (Narikawa et al., 2008; Rockwell et al., 2008), and while the mechanism of their photocycle and spectral diversity is being studied to some extent (Freer et al., 2012; Rockwell et al., 2012a; Rockwell et al., 2012b), additional characterizations of these proteins will contribute significantly to our knowledge of phytochrome structure, function and evolution.
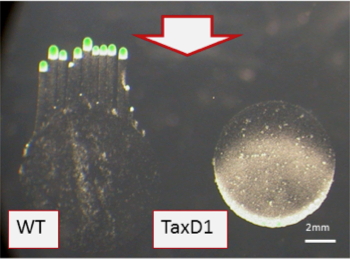
Figure 1. Synechocystis PCC6803 Type IV pilus-dependent motility assay on soft agarose plates, with directional white light (shown by arrow). The cells exhibit positive phototaxis, with finger like projections moving toward the light (left), while a mutant in the CBCR-type photoreceptor (TaxD1) loses positive phototaxis (right).
In conclusion, cyanobacteria contain active photosensors that have a single GAF domain, representing some of the smallest identified photosensory modules. However, there is also a bewildering multitude of cyanobacterial proteins that have a combination of GAF, PHY and PAS domains (Auldridge and Forest, 2011; Ulijasz et al., 2009). These domains can bind different chromophores, so their ability to absorb different wavelengths of light can also be modulated. Finally, these photosensory modules can be attached to a variety of output domains, such as histidine kinases, methyl accepting domains, response regulators, and GGDEF-EAL domains. This strategy of mixing and matching photosensory and output modules provides cyanobacteria with a diversity of proteins that can respond to environmental signals. The challenge for the future is establishing how these multiple photoswitches work together in a cell to optimize growth and modulate behavior under varying light conditions.
III. COMPLEMENTARY CHROMATIC ADAPTATION
A. The Phycobilisome (PBS)
The vivid coloration of cyanobacteria is mostly a consequence of the presence of the PBS, a light harvesting complex that is peripheral to the photosynthetic membranes, and in some organisms (and under some conditions) accounts for ~30% of total cellular protein (Grossman et al., 1995; Tandeau de Marsac and Houmard, 1993). This light harvesting complex absorbs excitation energy and efficiently transfers the absorbed energy to photosynthetic reaction centers (Porter et al., 1978; Searle et al., 1978). PBS levels in cyanobacterial cells may undergo extreme variation, especially as light and nutrient availability in the environment fluctuate. Numerous studies on the ways in which PBS levels change in response to macronutrient deprivation (Collier and Grossman, 1992, 1994; Kato et al., 2011; Sato et al., 2008), or are modified in response to the prevalent wavelengths of light in the environment in the process of CA, have been carried out by our group and other groups (Grossman, 2003; Grossman and Kehoe, 1997; Gutu and Kehoe, 2012; Kehoe, 2010; Tandeau de Marsac and Houmard, 1993). Before exploring CA in greater detail, we will describe key features of the PBS.
Gantt and Conti performed pioneering work that led to the first isolation and description of the PBS (Gantt and Conti, 1966a, b), which are present in both cyanobacteria and red algae. The PBS appear as large, knob-like structures that bulge from the surface of the photosynthetic membranes; these membranes, called thylakoids, are dominant features in the cytoplasm of cyanobacteria, often forming concentric circles around the cell periphery. Each PBS is composed of two domains called the rods and core. Each of these domains contain pigmented phycobiliproteins, which absorb excitation energy, and nonpigmented linker polypeptides, which are important structural components of the PBS.
The rods can be composed of the pigmented phycobiliproteins, phycocyanin (PC, absorption max ~620 nm) and phycoerythrin (PE, absorption maximum ~540 nm), although many cyanobacteria have PBS that are exclusively composed of PC. The core contains allophycocyanin (AP, absorption maximum ~620 nm). Pigmentation of the phycobiliproteins is a consequence of covalent binding of apo-phycobiliproteins to linear tetrapyrrole chromophores (e.g., phycocyanobilin, phycourobilin or phycoerythrobilin). Each phycobiliprotein is made up of a specific α and β subunit, which assemble into heterodimers (these heterodimers are called phycobiliprotein ‘monomers’), and the monomers then assemble into trimers and hexamers, forming structures that resemble a donut (phycobiliprotein disks).
Specific lyases catalyze the covalent bonding of the chromophore to the apophycobiliprotein (Biswas et al., 2011; Fairchild and Glazer, 1994; Fairchild et al., 1992; Kahn et al., 1997). The nonpigmented linker polypeptides serve as structural elements of the PBS. They are critical for assembling phycobiliprotein arrays into rod and core substructures, stabilizing these substructures, and conferring individual trimeric and hexameric phycobiliprotein disks with spatial and biophysical features that create a highly efficient light harvesting complex. The linker polypeptides facilitate efficient, unidirectional flow of excitation energy from the periphery of the rod substructures to the core of the complex, and then to the photosynthetic reactions centers.
Many investigators have performed critical experiments to elucidate the structure and function of PBS; these include Elizabeth Gantt (Dilworth and Gantt, 1981; Gantt et al., 1979; Gantt et al., 1976a; Gantt et al., 1976b; Redlinger and Gantt, 1981), Alexander Glazer (Glazer, 1982, 1985; Glazer and Clark, 1986; Glazer et al., 1983), Herbert Zuber (Glauser et al., 1992, Glauser, 1993 #1204) and Robert Huber (Schirmer et al., 1987; Schirmer et al., 1986). Detailed descriptions of the PBS structure, mostly of the hemidiscoidal type, can be found in a number of review articles (Adir, 2005; Bryant et al., 1979; Gantt, 1980; Glazer, 1985; Glazer et al., 1983; Sidler, 1994). Recent work describing the regulation of PBS biosynthesis as it relates to nutrient availability and light quality are mentioned below. In the remainder of this article, we focus on the ways in which light quality modulates PBS structure and function in the process of CA, and the regulatory elements that are critical for this process.
B. The Initial Observations
Well over a hundred years ago it was noted that certain cyanobacteria were able to change their pigmentation when the wavelengths of light to which they were exposed was altered (Engelmann, 1902); this phenomenon was later designated chromatic adaptation or CA (Gaidukov, 1903). The ability to modulate pigment content has been observed in cyanobacteria growing in a number of different environments (Acinas et al., 2009; Dufresne et al., 2008; Duxbury et al., 2009; Postius et al., 2001; Tandeau de Marsac, 1977, 1983; Tandeau de Marsac et al., 1980), allowing these organisms to efficiently absorb prevalent wavelengths of excitation energy in their immediate surroundings. This may be important in aqueous environments where certain wavelengths penetrate a water column better than others (e.g., BL penetrates deeper than RL). Light quality can also cause changes in the morphology of cyanobacteria. For example, it has been noted that hormogonia (cell filaments by which cyanobacteria propagate) formation and the morphology of F. diplosiphon cells are impacted by light quality (Bennett and Bogorad, 1973; Bordowitz and Montgomery, 2008; Damerval et al., 1991), with larger, more rounded cells appearing when cultures are maintained in RL compared to GL (Bennett and Bogorad, 1973; Bordowitz and Montgomery, 2008).
Additionally, gas vesicle formation can be impacted by light quality (Damerval et al., 1991; Tandeau de Marsac et al., 1988). Some of these developmental changes may impact light utilization as well as the position of the cyanobacteria with respect to the light gradient. Furthermore, while some morphological features noted above appear to be controlled by regulators of CA, others are not (Bordowitz and Montgomery, 2008; Bordowitz et al., 2010; Pattanaik et al., 2011). Finally, it was recently suggested that light-triggered changes in cell shape might involve reactive oxygen species (Singh and Montgomery, 2012).
CA responses observed in cyanobacteria have been categorized into three groups. Group I cyanobacteria do not alter the components of the PBS in response to light quality. In group II organisms, the level of PE increases in GL relative to RL, while PC levels do not respond to these light qualities. In group III organisms, the PE levels are high in GL and low in RL while the PC levels are high in RL and low in GL. The phenomenon observed in this last group of organisms has been termed complementary chromatic adaptation (CCA) since cell pigmentation adjusts in a way that is complementary to the wavelengths of light in which the organism is being grown (Figure 2). The CA group I, II and III organisms are also called type I or CA1 species, type II or CA2 species and type III or CA3 species. In another mode of CA (sometimes called CA4), the specific chromophore that is attached to some of the phycobiliproteins may change, which alters the absorbance of the PBS.
For example, in some strains of Synechococcus, PE subunits can associate with phycourobilin when the cells are exposed to BL, and with phycoerythrobilin when the cells are exposed to GL (Everroad et al., 2006; Palenik, 2001); the former better absorbs BL (peak absorbance ~495 nm) while the later better absorb GL (peak absorbance ~540 nm).
The remainder of this article focuses on CCA or CA3. As shown in Figure 2, CCA is visually dramatic, with marked differences in pigmentation of the filaments of F. diplosiphon (also called Calothrix and Tolypothrix sp. PCC7601) after growth of the cells on solid medium in RL compared to GL. The ability to perform CA3 may confer an advantage to cyanobacteria as the color of the light in the environment changes (Stomp et al., 2004; Stomp et al., 2008), although more studies of organisms growing in their natural environments may be required to detail the advantages associated with CCA-competent cells.
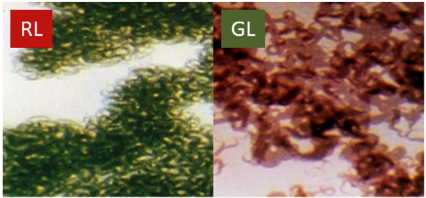
Figure 2. Complementary chromatic adaptation in the filamentous cyanobacterium Fremyella diplosiphon during growth in RL and GL. Left: F. diplosiphon cells grown in RL. Right: F. diplosiphon cells grown in GL. The visually noticeable differences in cell pigmentation represent differences in the specific phycobiliproteins that are associated with the PBS (see Figure 3).
C. The Phenomenon of CCA
In early studies, action spectra associated with control of PC and PE synthesis, and the changes in chromophore composition of the PBS were established for two organisms that perform CCA, T. tenuis and F. diplosiphon (Bennett and Bogorad, 1971, 1973; Bogorad, 1975; Diakoff and Scheibe, 1973; Haury and Bogorad, 1977; Vogelmann and Scheibe, 1978). When cells were exposed to GL of ~550 nm, PE accumulation was high while PC accumulation was low. In contrast, RL of ~640 nm led to high accumulation of PC and low accumulation of PE. Changes in the protein-pigment structure of the PBS as a consequence of exposure to different qualities of light are depicted in Figure 3. This spectral and biochemical data suggest that the process of CCA is under the control of a photoreceptor(s) that absorbs both RL and GL, but that it elicits different responses in its RL- and GL-absorbing forms. The synthesis and assembly of PC into PBS are favored when the cells are absorbing RL, while the synthesis and assembly of PE are favored when the cells are absorbing GL. The mixture of wavelengths in natural sunlight would elicit the biosynthesis of PBS with intermediate PC and PE levels.
As molecular technology grew more sophisticated, the genes encoding polypeptides that form the structure of the PBS were characterized [summarized in (Conley et al., 1985; Conley et al., 1986; Grossman et al., 1995; Houmard et al., 1988; Tandeau de Marsac and Houmard, 1993)], yielding complete, deduced amino acid sequences for both phycobiliprotein and linker polypeptides, which in turn has helped establish molecular details associated with the PBS structure. Molecular information has also enabled new ways of examining the regulation of genes encoding individual linker and phycobiliprotein polypeptides, and especially the role of light quality in the control of gene expression.
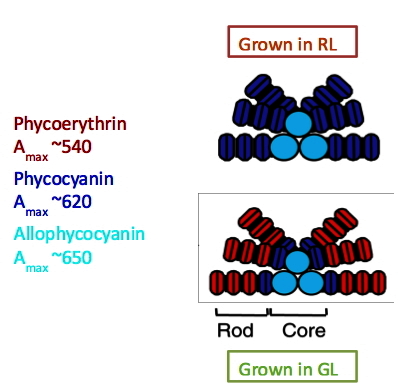
Figure 3. Dramatic changes in PBS composition after exposure of cyanobacterial cells that perform CCA to different wavelengths of light (e.g., RL of ~620 nm and GL of ~540 nm). The top panel represents a PBS from cells grown in RL while the bottom panel represents a PBS from cells grown in GL. Here, the rods are shown to be composed of three (in RL) or four (in GL) phycobiliprotein disks (hexamers), with the pigmented protein in the rods in RL being almost exclusively PC (blue color), and the pigmented protein in the rods in GL being mostly PE (red color, with PC making up the disk most proximal to the core). The linker polypeptide components of the PBS also exhibit changes, although this is not shown in this figure.
Genes encoding phycobiliprotein and linker polypeptides have now been cloned from many organisms. F. diplosiphon has one set of genes for α (cpeA) and β (cpeB) subunits of PE (cpeBA operon). A separate operon (cpeCDESTR) encodes the PE linkers (cpeCDE), associated lyases that likely attach the chromophores to PE (cpeST), and a regulatory element (cpeR; see below). F. diplosiphon has three sets of genes for α (cpcA) and β (cpcB) subunits of PC, which are in the operons cpcB1A1, cpcB2A2 and cpcB3A3. The cpcB1A1 genes are constitutively expressed (Conley et al., 1988; Conley et al., 1986; Houmard et al., 1988; Mazel et al., 1988), with the monomeric phycobiliprotein product designated PCc (or PC1). The cpcB2A2 genes are induced in RL (Conley et al., 1988; Conley et al., 1985; Lomax et al., 1987), with the monomeric phycobiliprotein product designated PCi (or PC2). The cpcB3A3 genes are expressed when sulfur becomes limiting in the environment, with the monomeric phycobiliprotein product designated PCs (or PC3) (Mazel and Marliere, 1989). Downstream of cpcB2A2 are the genes cpcHID, which encode the three linker polypeptides that bind PCi hexamers; these genes are RL-inducible and co-transcribed with cpcB2A2 as part of the cpcB2A2HID operon (Lomax et al., 1987).
Differences in levels of phycobiliproteins in the different light qualities are a consequence of altered transcription from cpeBA, cpeCDESTR and cpcB2A2HID (Casey and Grossman, 1994; Conley et al., 1985; Mazel et al., 1986; Oelmuller et al., 1988a; Oelmuller et al., 1988b). Expression of genes encoding PE, and the PE-associated linker, ligase and regulatory polypeptides of the cpeCDESTR operon, are also under the control of RL and GL (transcribed in GL) (Federspiel and Grossman, 1990; Federspiel and Scott, 1992; Mazel et al., 1986). Furthermore, the genes encoding enzymes required for the synthesis of the chromophores that associate with the different phycobiliproteins are also regulated by RL and GL (Alvey et al., 2007; Alvey et al., 2003).
D. Dissection of CCA Through the Isolation and Analysis of Mutants
The ability to visually screen for pigment mutants that exhibit aberrant CCA has been a powerful way to identify regulatory elements in this process in F. diplosiphon (Balabas et al., 2003; Bruns et al., 1989; Casey et al., 1997; Chiang et al., 1992; Cobley and Miranda, 1983; Kehoe and Grossman, 1996, 1997; Li and Kehoe, 2005; Tandeau de Marsac, 1983). Characterization of the genes altered in the mutant strains, coupled with complementation of the mutant phenotypes, has led to identification of a series of regulatory elements critical for different aspects of CCA. Some of the first mutants analyzed were designated FdR. These mutants maintain red pigmentation in both RL and GL, and cannot activate the cpc2 genes in RL.
The lesions in these strains are in genes designated rcaC and rcaF (Chiang et al., 1992; Grossman and Kehoe, 1997; Kehoe and Grossman, 1996, 1997), which encode response regulators associated with two component regulatory systems [for reviews see (Galperin et al., 2001; Marijuan et al., 2010; Mitrophanov and Groisman, 2008)]. RcaF is a small response regulator that has a receiver domain but no identifiable output domain (Kehoe and Grossman, 1997). RcaC has an OmpR-type DNA-binding motif and characteristic domains with potential phosphorylation sites which include N- and C-terminal aspartate residues (D51 and D576, respectively), as well as a histidine residue (H316) that is part of an H block domain (often present in sensors). Both D51 and H316 residues of RcaC have been shown to be critical for CCA (Kehoe and Grossman, 1995; Li and Kehoe, 2005).
Another characterized CCA mutant has been designated FdBk. This strain appears purple-black as a consequence of having both PC and PE when the cells are exposed to either RL or GL. The gene disrupted in this mutant has been designated RcaE. This gene encodes a protein of 74 kDa (Kehoe and Grossman, 1996), has C-terminal motifs required for histidine kinase activity (N, G1, F and G2), an H block and an N terminus with a tetrapyrrole chromophore attachment domain, similar to the GAF domain associated with phytochrome (including the cysteine residue that binds the chromophore).
Thus, this protein is considered a CBCR, and was one of the first to be functionally identified. Biochemical and genetic analyses have shown that RcaE is required for both the RL and GL responses in F. diplosiphon (Terauchi et al., 2004). Following the discovery of RcaE, many other phytochrome-like photoreceptors of the CBCR type (no PAS or Phy domains) have been identified in cyanobacteria, as discussed above. Finally, yet another regulatory element, the CpeR element, encoded by the terminal gene of the cpeCDESTR operon, is part of the Cgi (Control of green light induction) regulatory system and is required for activation of both cpeBA and pebAB operons in GL (Alvey et al., 2003; Cobley et al., 2002; Seib and Kehoe, 2002).
Rca control of the RL-inducible PC genes, and Rca and Cgi control of the PE genes are linked by the regulatory element RcaC; this link is depicted in the model presented in Figure 4. Light absorbed by the RcaE CBCR photoreceptor initiates a phosphorelay (a multi-stage process, involving the movement of phosphoryl groups through the activity of protein kinases and phosphatases) yielding phosphorylated forms of RcaF and RcaC. Phosphorylated RcaC binds to specific promoter motifs that have been designated the L box. These motifs are present upstream of the cpcB2A2HID, pcyA and cpeCDESTR operons (Li et al., 2008), but are not upstream of the cpeBA operon (which is not active in RL). RcaC binding activates the cpcB2A2HID and pcyA operons in RL, while at the same time binding to and repressing the activity of the cpeCDESTR operon.
These regulatory events in RL promote the synthesis of PBS with high levels of PC, but very low levels of PE. Conversely, in GL, RcaC is not phosphorylated (and may function as a phosphatase) and cannot activate the cpcB2A2HID or pcyA operons. This unphosphorylated DNA binding protein also no longer represses transcription of the cpeCDESTR operon. Activation of this operon leads to the synthesis of the PE-associated linker polypeptides (CpeCDE) and the lyases (CpeST) that attach the chromophore to PE, as well as the CpeR regulatory protein. CpeR then activates the cpeBA and the pebAB genes; the former encodes the PE α and β subunits, while the latter encode proteins involved in phycoerythrobilin biosynthesis. These events in GL lead to the synthesis of a PBS with no PCi (they will still have the PCc subunits that make up the PC hexamers immediately contiguous to the PBS core) and high levels of PE hexamers that form most of the rod substructure. A more detailed discussion of regulatory events that control CCA are discussed by Gutu and Kehoe (Gutu and Kehoe, 2012).
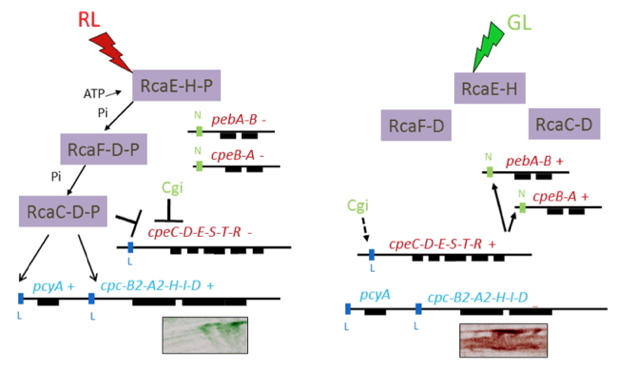
Figure 4. Depiction of CCA in Fremyella diplosiphon. Events accompanying exposure of cells to RL are shown on the left, and to GL are shown on the right. (Left) RcaE absorbs RL, undergoes autophosphorylation and initiates a phosphotransfer to RcaF and then to RcaC. The phosphoryl group (P) is attached to a histidine (H) in RcaE, and to aspartates (D) in RcaF and RcaC. Phosphorylated RcaC binds to the L boxes, activating transcription from the cpc2 operon (cpcB2A2HID) and pcyA gene (the PcyA protein is involved in phycocyanobilin synthesis) and repressing transcription from the cpeCDESTR operon (along with the Cgi system). These events lead to the synthesis of α and β subunits of PC2 or PCi along with the associated linker polypeptides. The cpeCDESTR, pebAB and cpeBA genes are not active under these conditions, so no PE is synthesized. (Right) RcaE absorbs GL, which prevents the phosphorelay, so RcaE, RcaF and RcaC remain unphosphorylated; under these conditions RcaE may function as a phosphatase. The cpeCDESTR genes are no longer repressed by RcaC (and possibly also somewhat activated by the Cgi system, as indicated by a broken line) and the regulatory element CpeR is synthesized. This protein binds to the N boxes of the pebA and cpeBA operon, causing gene activation, which leads to the synthesis of PE. The cells growing on agar plates are shown below the cpc2 operon; on the left are RL grown cells, and on the right are GL grown cells.
CONCLUDING REMARKS
Chromatic adaptation (CA) is a complex, highly regulated process that appears to be tailored in specific cyanobacteria. It reflects a diversity of mechanisms that modulate the absorption properties of the light harvesting phycobilisome (PBS) in cyanobacteria. In some cases there is strong differential expression of genes encoding phycobiliprotein subunits and linker polypeptides as the wavelengths of light in the environment change, while in other cases light quality may elicit changes in the specific chromophores that are attached to phycobiliproteins. The consequence of these processes is a PBS that is most efficient in the capture of the available light. While we have made considerable progress in defining elements that regulate CA, other elements remain to be investigated. Environmental features such as light intensity, nutrient availability, temperature and biotic interactions impact the biogenesis of PBS, providing inputs that tune its final levels and pigment-protein composition.
A strong impact of reduced nutrient availability on PBS biogenesis has been noted (Collier and Grossman, 1992, 1994; Grossman et al., 1993a; Richaud et al., 2001), as well as regulatory factors that control expression from cpc1 (Kahn and Schaefer, 1997; Manna et al., 2000), which encodes the putative constitutive phycocyanin (PC) subunits. Furthermore, while one protein factor that binds to cpeBA and regulates its expression (Li et al., 2008) has been identified, there is evidence that additional elements may interact with the promoter of this operon, and that some of these elements may be affected by post-translational modifications (Sobczyk et al., 1993). Finally, since light harvesting function impacts photosynthetic outputs, it is likely that the products of photosynthesis will participate in feedback loops that tune light harvesting function and biogenesis. Such outputs might include specific metabolites, redox carriers and regulators, and levels of reactive oxygen species.
REFERENCES
Acinas, S.G., Haverkamp, T.H., Huisman, J., Stal, L.J., 2009. Phenotypic and genetic diversification of Pseudanabaena spp. (cyanobacteria). ISME J 3, 31-46.
Adir, N., 2005. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosyn Res 85, 15-32.
Alvey, R.M., Bezy, R.P., Frankenberg-Dinkel, N., Kehoe, D.M., 2007. A light regulated OmpR-class promoter element co-ordinates light-harvesting protein and chromophore biosynthetic enzyme gene expression. Mol Microbiol 64, 319-332.
Alvey, R.M., Karty, J.A., Roos, E., Reilly, J.P., Kehoe, D.M., 2003. Lesions in phycoerythrin chromophore biosynthesis in Fremyella diplosiphon reveal coordinated light regulation of apoprotein and pigment biosynthetic enzyme gene expression. Plant Cell 15, 2448-2463.
Anders, K., von Stetten, D., Mailliet, J., Kiontke, S., Sineshchekov, V.A., Hildebrandt, P., Hughes, J., Essen, L.O., 2011. Spectroscopic and photochemical characterization of the red-light sensitive photosensory module of Cph2 from Synechocystis PCC 6803. Photochem Photobiol 87, 160-173.
Auldridge, M.E., Forest, K.T., 2011. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol 46, 67-88.
Balabas, B.E., Montgomery, B.L., Ong, L.E., Kehoe, D.M., 2003. CotB is essential for complete activation of green light-induced genes during complementary chromatic adaptation in Fremyella diplosiphon. Mol Microbiol 50, 781-793.
Bennett, A., Bogorad, L., 1971. Properties of subunits and aggregates of blue-green algal biliproteins. Biochem 10, 3625-3634.
Bennett, A., Bogorad, L., 1973. Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol 58, 419-435.
Bhaya, D., Takahashi, A., Grossman, A.R., 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci USA 98, 7540-7545.
Biswas, A., Boutaghou, M.N., Alvey, R.M., Kronfel, C.M., Cole, R.B., Bryant, D.A., Schluchter, W.M., 2011. Characterization of the activities of the CpeY, CpeZ, and CpeS bilin lyases in phycoerythrin biosynthesis in Fremyella diplosiphon strain UTEX 481. J Biol Chem 286, 35509-35521.
Bogorad, L., 1975. Phycobiliproteins and complementary chromatic adaptation. Annu Rev Plant Physiol 26, 369-401.
Bordowitz, J.R., Montgomery, B.L., 2008. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J Bacteriol 190, 4069-4074.
Bordowitz, J.R., Whitaker, M.J., Montgomery, B.L., 2010. Independence and interdependence of the photoregulation of pigmentation and development in Fremyella diplosiphon. Communicative & Integrative Biology 3, 151-153.
Bruns, B.U., Briggs, W.R., Grossman, A.R., 1989. Molecular characterization of phycobilisome regulatory mutants of Fremyella diplosiphon. J Bacteriol 171, 901-908.
Bryant, D., Guglielmi, G., Tandeau de Marsac, N., Castets, A.M., Cohen-Bazire, G., 1979. The structure of cyanobacterial phycobilisomes. Arch Microbiol 123, 113-127.
Casal, J.J., 2012. Shade avoidance. The Arabidopsis book / American Society of Plant Biologists 10, e0157.
Casey, E.S., Grossman, A., 1994. In vivo and in vitro characterization of the light-regulated cpcB2A2 promoter of Fremyella diplosiphon. J Bacteriol 176, 6362-6374.
Casey, E.S., Kehoe, D.M., Grossman, A.R., 1997. Suppression of mutants aberrant in light intensity responses of complementary chromatic adaptation. J Bacteriol 179, 4599-4606.
Chen, M., Chory, J., 2011. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21, 664-671.
Chiang, G.G., Schaefer, M.R., Grossman, A.R., 1992. Complementation of a red-light-indifferent cyanobacterial mutant. Proc Natl Acad Sci USA 89, 9415-9419.
Cobley, J.G., Clark, A.C., Weerasurya, S., Queseda, F.A., Xiao, J.Y., Bandrapali, N., D'Silva, I., Thounaojam, M., Oda, J.F., Sumiyoshi, T., Chu, M.H., 2002. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR). Mol Microbiol 44, 1517-1531.
Cobley, J.G., Miranda, R.D., 1983. Mutations affecting chromatic adaptation in the cyanobacterium Fremyella diplosiphon. J Bacteriol 153, 1486-1492.
Collier, J.L., Grossman, A.R., 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol 174, 4718-4726.
Collier, J.L., Grossman, A.R., 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J 13, 1039-1047.
Conley, P.B., Lemaux, P.G., Grossman, A., 1988. Molecular characterization and evolution of sequences encoding light-harvesting components in the chromatically adapting cyanobacterium Fremyella diplosiphon. J Mol Biol 199, 447-465.
Conley, P.B., Lemaux, P.G., Grossman, A.R., 1985. Cyanobacterial light-harvesting complex subunits encoded in two red light-induced transcripts. Science 230, 550-553.
Conley, P.B., Lemaux, P.G., Lomax, T.L., Grossman, A.R., 1986. Genes encoding major light-harvesting polypeptides are clustered on the genome of the cyanobacterium Fremyella diplosiphon. Proc Natl Acad Sci USA 83, 3924-3928.
Damerval, T., Guglielmi, G., Houmard, J., De Marsac, N.T., 1991. Hormogonium differentiation in the cyanobacterium Calothrix: A photoregulated developmental process. Plant Cell 3, 191-201.
Davis, S.J., Vener, A.V., Vierstra, R.D., 1999. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286, 2517-2520.
Diakoff, I., Scheibe, J., 1973. Action spectrum for chromatic adaptation in Tolypothrix tenuis. Plant Physiol 51, 382-385.
Dilworth, M.F., Gantt, E., 1981. Phycobilisome-thylakoid topography on photosynthetically active vesicles of Porphyridium cruentum. Plant Physiol 67, 608-612.
Dufresne, A., Ostrowski, M., Scanlan, D.J., Garczarek, L., Mazard, S., Palenik, B.P., Paulsen, I.T., de Marsac, N.T., Wincker, P., Dossat, C., Ferriera, S., Johnson, J., Post, A.F., Hess, W.R., Partensky, F., 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biology 9, R90.
Durnford, D.G., Falkowski, P.G., 1997. Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosyn Res 53, 229–241.
Duxbury, Z., Schliep, M., Ritchie, R.J., Larkum, A.W., Chen, M., 2009. Chromatic photoacclimation extends utilisable photosynthetically active radiation in the chlorophyll d-containing cyanobacterium, Acaryochloris marina. Photosyn Res 101, 69-75.
Eberhard, S., Finazzi, G., Wollman, F.A., 2008. The dynamics of photosynthesis. Annu Rev Genet 42, 463-515.
Engelmann, T.W., 1902. Untersuchungen über die qualitativen Beziehungen zwieschen Absorbtion des Lichtes und Assimilation in Pflanzenzellen. I. Das Mikrospectraphotometer, ein Apparat zur qualitativen Mikrospectralanalyse. II. Experimenttelle Grundlangen zur Ermittelung der quantitativen Beziehungen zwieschen Assimilationsenergie und Absorptiongrösse. III. Bestimmung der Vertheilung der Energie im Spectrum von Sonnenlicht mittel Bacterien-Methode und quantitativen Mikrospectralanalyse. Bot Z 42, 81-105.
Escoubas, J.M., Lomas, M., LaRoche, J., Falkowski, P.G., 1995. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92, 10237-10241.
Everroad, C., Six, C., Partensky, F., Thomas, J.C., Holtzendorff, J., Wood, A.M., 2006. Biochemical bases of type IV chromatic adaptation in marine Synechococcus spp. J Bacteriol 188, 3345-3356.
Fairchild, C.D., Glazer, A.N., 1994. Nonenzymatic bilin addition to the alpha subunit of an apophycoerythrin. J Biol Chem 269, 28988-28996.
Fairchild, C.D., Zhao, J., Zhou, J., Colson, S.E., Bryant, D.A., Glazer, A.N., 1992. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc Natl Acad Sci USA 89, 7017-7021.
Federspiel, N.A., Grossman, A.R., 1990. Characterization of the light-regulated operon encoding the phycoerythrin-associated linker proteins from the cyanobacterium Fremyella diplosiphon. J Bacteriol 172, 4072-4081.
Federspiel, N.A., Scott, L., 1992. Characterization of a light-regulated gene encoding a new phycoerythrin-associated linker protein from the cyanobacterium Fremyella diplosiphon. J Bacteriol 174, 5994-5998.
Fiedler, B., Borner, T., Wilde, A., 2005. Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: role of different photoreceptors. Photochem Photobiol 81, 1481-1488.
Frankhauser, C., Casal, J.J., 2004. Phenotypic characterization of a photomorphogenic mutant. Plant J 39, 747-760.
Franklin, K.A., 2008. Shade avoidance. The New Phytol 179, 930-944.
Franklin, K.A., Quail, P.H., 2010. Phytochrome functions in Arabidopsis development. J Exp Bot 61, 11-24.
Freer, L.H., Kim, P.W., Corley, S.C., Rockwell, N.C., Zhao, L., Thibert, A.J., Lagarias, J.C., Larsen, D.S., 2012. Chemical inhomogeneity in the ultrafast dynamics of the DXCF cyanobacteriochrome Tlr0924. J Phys Chem B 116, 10571-10581.
Fujita, H., Murakami, A., Aizawa, K., 1994. Short term and long-term adaptation of the photosynthetic apparatus; homeostatic properties of thylakoids, in: Bryant, D. (Ed.), The Molecular Biology of Cyanobacteria. Kluwer Academic Publisher, Dordrecht, pp. 677-692.
Gaidukov, N., 1903. Die Farbenveränderung bei den Prozessen der komplementären chromatischen Adaptation. Der Deutsch Bot Ges 21, 517-522.
Galperin, M.Y., Nikolskaya, A.N., Koonin, E.V., 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203, 11-21.
Gantt, E., 1980. Structure and function of phycobilisomes: light-harvesting pigment complex in red and blue-green algae. Int Rev Cyt 66, 45-80.
Gantt, E., Conti, S.F., 1966a. Granules associated with the chloroplast Lamellae of Porphyridium cruentum. J Cell Biol 29, 423-434.
Gantt, E., Conti, S.F., 1966b. Phycobiliprotein localization in algae, Brookhaven Symp Biol - Energy Conversion by the Photosynthetic Apparatus, New York, pp. 393-405.
Gantt, E., Lipschultz, C.A., Grabowski, J., Zimmerman, B.K., 1979. Phycobilisomes from blue-green and red algae: isolation criteria and dissociation characteristics. Plant Physiol 63, 615-620.
Gantt, E., Lipschultz, C.A., Zilinskas, B., 1976a. Further evidence for a phycobilisome model from selective dissociation, fluorescence emission, immunoprecipitation, and electron microscopy. Biochim Biphys Acta 430, 375-388.
Gantt, E., Lipschultz, C.A., Zilinskas, B.A., 1976b. Phycobilisomes in relation to the thylakoid membranes. Brookhaven Symposia in Biology, 347-357.
Glauser, M., Bryant, D.A., Frank, G., Wehrli, E., Rusconi, S.S., Sidler, W., Zuber, H., 1992. Phycobilisome structure in the cyanobacteria Mastigocladus laminosus and Anabaena sp. PCC 7120. E J Biochem / FEBS 205, 907-915.
Glazer, A.N., 1982. Phycobilisomes: structure and dynamics. Annual review of microbiology 36, 173-198.
Glazer, A.N., 1985. Light harvesting by phycobilisomes. Annu Rev Biphys Biophys Chem 14, 47-77.
Glazer, A.N., Clark, J.H., 1986. Phycobilisomes: macromolecular structure and energy flow dynamics. Biophys J 49, 115-116.
Glazer, A.N., Lundell, D.J., Yamanaka, G., Williams, R.C., 1983. The structure of a "simple" phycobilisome. Annales de Microbiologie 134B, 159-180.
Grossman, A.R., 2003. A molecular understanding of complementary chromatic adaptation. Photosyn Res 76, 207-215.
Grossman, A.R., D., B., Apt, K.E., Kehoe, D.M., 1995. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu Rev Genet 29, 231–288.
Grossman, A.R., Kehoe, D.M., 1997. Phosphorelay control of phycobilisome biogenesis during complementary chromatic adaptation. Photosyn Res 53, 95-108.
Grossman, A.R., Schaefer, M.R., Chiang, G.G., Collier, J.L., 1993a. Environmental effects on the light-harvesting complex of cyanobacteria. J Bacteriol 175, 575-582.
Grossman, A.R., Schaefer, M.R., Chiang, G.G., Collier, J.L., 1993b. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev 57, 725-749.
Gutu, A., Kehoe, D.M., 2012. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant 5, 1-13.
Haury, J.F., Bogorad, L., 1977. Action spectra for phycobiliprotein synthesis in a chromatically adapting cyanophyte, Fremyella diplosiphon. Plant Physiol 60, 835-839.
Houmard, J., Capuano, V., Coursin, T., Tandeau de Marsac, N., 1988. Genes encoding core components of the phycobilisome in the cyanobacterium Calothrix sp. strain PCC 7601: occurrence of a multigene family. J Bacteriol 170, 5512-5521.
Hughes, J., 2010. Phytochrome three-dimensional structures and functions. Biochem Soc Trans 38, 710-716.
Ikeuchi, M., Ishizuka, T., 2008. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol 7, 1159-1167.
Ishizuka, T., Shimada, T., Okajima, K., Yoshihara, S., Ochiai, Y., Katayama, M., Ikeuchi, M., 2006. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant & Cell Phys 47, 1251-1261.
Kahn, K., Mazel, D., Houmard, J., Tandeau de Marsac, N., Schaefer, M.R., 1997. A role for cpeYZ in cyanobacterial phycoerythrin biosynthesis. J Bacteriol 179, 998-1006.
Kahn, K., Schaefer, M.R., 1997. RpbA controls transcription of the constitutive phycocyanin gene set in Fremyella diplosiphon. J Bacteriol 179, 7695-7704.
Kami, C., Lorrain, S., Hornitschek, P., Fankhauser, C., 2010. Light-regulated plant growth and development. Curr Topics Devel Biol 91, 29-66.
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., Kimura, T., Hosouchi, T., Matsuno, A., Muraki, A., Nakazaki, N., Naruo, K., Okumura, S., Shimpo, S., Takeuchi, C., Wada, T., Watanabe, A., Yamada, M., Yasuda, M., Tabata, S., 1996a. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3, 109-136.
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., Kimura, T., Hosouchi, T., Matsuno, A., Muraki, A., Nakazaki, N., Naruo, K., Okumura, S., Shimpo, S., Takeuchi, C., Wada, T., Watanabe, A., Yamada, M., Yasuda, M., Tabata, S., 1996b. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement). DNA Res 3, 185-209.
Kato, H., Kubo, T., Hayashi, M., Kobayashi, I., Yagasaki, T., Chibazakura, T., Watanabe, S., Yoshikawa, H., 2011. Interactions between histidine kinase NblS and the response regulators RpaB and SrrA are involved in the bleaching process of the cyanobacterium Synechococcus elongatus PCC 7942. Plant & Cell Physiol 52, 2115-2122.
Kehoe, D., Grossman, A.R., 1995. The use of site directed mutagenesis in the analysis of complementary chromatic adaptation, in: Mathis, P. (Ed.), Proceedings from the Xth International Photosynthesis Congress; Photosynthesis: from Light to Biosphere. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 501-504.
Kehoe, D.M., 2010. Chromatic adaptation and the evolution of light color sensing in cyanobacteria. Proc Natl Acad Sci USA 107, 9029-9030.
Kehoe, D.M., Grossman, A.R., 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273, 1409-1412.
Kehoe, D.M., Grossman, A.R., 1997. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J Bacteriol 179, 3914-3921.
Li, L., Alvey, R.M., Bezy, R.P., Kehoe, D.M., 2008. Inverse transcriptional activities during complementary chromatic adaptation are controlled by the response regulator RcaC binding to red and green light-responsive promoters. Mol Microbiol 68, 286-297.
Li, L., Kehoe, D.M., 2005. In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol Microbiol 55, 1538-1552.
Lomax, T.L., Conley, P.B., Schilling, J., Grossman, A.R., 1987. Isolation and characterization of light-regulated phycobilisome linker polypeptide genes and their transcription as a polycistronic mRNA. J Bacteriol 169, 2675-2684.
Manna, P., Nieder, R.P., Schaefer, M.R., 2000. DNA-Binding properties of the Fremyella diplosiphon RpbA repressor. J Bacteriol 182, 51-56.
Marijuan, P.C., Navarro, J., del Moral, R., 2010. On prokaryotic intelligence: strategies for sensing the environment. Bio Systems 99, 94-103.
Mazel, D., Guglielmi, G., Houmard, J., Sidler, W., Bryant, D.A., Tandeau de Marsac, N., 1986. Green light induces transcription of the phycoerythrin operon in the cyanobacterium Calothrix 7601. Nucleic Acids Res 14, 8279-8290.
Mazel, D., Houmard, J., Tandean de Marsac, N., 1988. A multigene family in Calothrix sp. PCC 7601 encodes phycocyanin, the major component of the cyanobacterial light harvesting antenna. Mol Gen Genetic 211, 296-304.
Mazel, D., Marliere, P., 1989. Adaptive eradication of methionine and cysteine from cyanobacterial light-harvesting proteins. Nature 341, 245-248.
McCormac, A.C., Terry, M.J., 2002. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32, 549-559.
Millar, A.J., 2004. Input signals to the plant circadian clock. J Exper Bot 55, 277-283.
Mitrophanov, A.Y., Groisman, E.A., 2008. Positive feedback in cellular control systems. BioEssays 30, 542-555.
Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., Quail, P.H., 2004. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101, 16091-16098.
Montgomery, B.L., 2007. Sensing the Light: Photoreceptive Systems and Signal Transduction in Cyanobacteria. Mol Microbiology 64, 16-27.
Montgomery, B.L., Lagarias, J.C., 2002. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci 7, 357-366.
Nagatani, A., 2010. Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13, 565-570.
Nagatani, A., Kay, S.A., Deak, M., Chua, N.H., Furuya, M., 1991. Rice type I phytochrome regulates hypocotyl elongation in transgenic tobacco seedlings. Proc Natl Acad Sci USA 88, 5207-5211.
Narikawa, R., Fukushima, Y., Ishizuka, T., Itoh, S., Ikeuchi, M., 2008. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J Mol Biol 380, 844-855.
Narikawa, R., Suzuki, F., Yoshihara, S., Higashi, S., Watanabe, M., Ikeuchi, M., 2011. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant & Cell Physiol 52, 2214-2224.
Oelmuller, R., Conley, P.B., Federspiel, N., Briggs, W.R., Grossman, A.R., 1988a. Changes in accumulation and synthesis of transcripts encoding phycobilisome components during acclimation of Fremyella diplosiphon to different light qualities. Plant Physiol 88, 1077-1083.
Oelmuller, R., Grossman, A.R., Briggs, W.R., 1988b. Photoreversibility of the effect of red and green light pulses on the accumulation in darkness of mRNAs coding for phycocyanin and phycoerythrin in Fremyella diplosiphon. Plant Physiol 88, 1084-1091.
Osugi, A., Itoh, H., Ikeda-Kawakatsu, K., Takano, M., Izawa, T., 2011. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 157, 1128-1137. Palenik, B., 2001. Chromatic adaptation in marine Synechococcus strains. Appl Environ Microbiol 67, 991-994.
Pattanaik, B., Whitaker, M.J., Montgomery, B.L., 2011. Convergence and divergence of the photoregulation of pigmentation and cellular morphology in Fremyella diplosiphon. Plant Signaling & Behavior 6, 2038-2041.
Porter, G., Tredwell, C.J., Searle, G.F.W., Barber, J., 1978. Picosecond time-resolved energy transfer in Porphyridium cruentum. Part I. In the intact alga. Biochem Biophys Acta 501, 232-245.
Postius, C., Neuschaefer-Rube, O., Haid, V., Böger, P., 2001. N2-fixation and complementary chromatic adaptation in non-heterocystous cyanobacteria from Lake Constance. FEMS Microbiol Ecol 37, 117-125.
Redlinger, T., Gantt, E., 1981. Phycobilisome structure of Porphyridium cruentum: POLYPEPTIDE COMPOSITION. Plant Physiol 68, 1375-1379.
Richaud, C., Zabulon, G., Joder, A., Thomas, J.C., 2001. Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J Bacteriol 183, 2989-2994.
Rockwell, N.C., Lagarias, J.C., 2006. The structure of phytochrome: a picture is worth a thousand spectra. Plant Cell 18, 4-14.
Rockwell, N.C., Lagarias, J.C., 2010. A brief history of phytochromes. Chemphyschem : Euro J Chem Phys and Phys Chem 11, 1172-1180.
Rockwell, N.C., Martin, S.S., Gulevich, A.G., Lagarias, J.C., 2012a. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochem 51, 1449-1463.
Rockwell, N.C., Martin, S.S., Lagarias, J.C., 2012b. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochem 51, 3576-3585.
Rockwell, N.C., Njuguna, S.L., Roberts, L., Castillo, E., Parson, V.L., Dwojak, S., Lagarias, J.C., Spiller, S.C., 2008. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochem 47, 7304-7316.
Rockwell, N.C., Su, Y.S., Lagarias, J.C., 2006. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57, 837-858.
Rohmer, T., Lang, C., Hughes, J., Essen, L.O., Gartner, W., Matysik, J., 2008. Light-induced chromophore activity and signal transduction in phytochromes observed by 13C and 15N magic-angle spinning NMR. Proc Natl Acad Sci USA 105, 15229-15234.
Rohmer, T., Strauss, H., Hughes, J., de Groot, H., Gartner, W., Schmieder, P., Matysik, J., 2006. 15N MAS NMR studies of cph1 phytochrome: Chromophore dynamics and intramolecular signal transduction. J Phys Chem B 110, 20580-20585.
Romling, U., Gomelsky, M., Galperin, M.Y., 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57, 629-639.
Rosler, J., Jaedicke, K., Zeidler, M., 2010. Cytoplasmic phytochrome action. Plant & Cell Physiol 51, 1248-1254.
Sato, H., Fujimori, T., Sonoike, K., 2008. sll1961 is a novel regulator of phycobilisome degradation during nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 582, 1093-1096.
Savakis, P., De Causmaecker, S., Angerer, V., Ruppert, U., Anders, K., Essen, L.O., Wilde, A., 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol 85, 239-251.
Schirmer, T., Bode, W., Huber, R., 1987. Refined three-dimensional structures of two cyanobacterial C-phycocyanins at 2.1 and 2.5 Å resolution. A common principle of phycobilin-protein interaction. J Mol Biol 196, 677-695.
Schirmer, T., Huber, R., Schneider, M., Bode, W., Miller, M., Hackert, M.L., 1986. Crystal structure analysis and refinement at 2.5 Å of hexameric C-phycocyanin from the cyanobacterium Agmenellum quadruplicatum. The molecular model and its implications for light-harvesting. J Mol Biol 188, 651-676.
Schopf, J.W., 2002. The fossil record tracing the roots of the cyanobacterial lineage, in: Whitton, B.A., Potts, M. (Eds.), The Ecology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht, London, Boston, pp. 12-35.
Searle, G.F., Barber, J., Porter, G., Tredwell, C.J., 1978. Picosecond time-resolved energy transfer in Porphyridium cruentum. Part II. In the isolated light harvesting complex (phycobilisomes). Biochim Biophys Acta 501, 246-256.
Seib, L.O., Kehoe, D.M., 2002. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J Bacteriol 184, 962-970.
Sellaro, R., Pacin, M., Casal, J.J., 2012. Diurnal dependence of growth responses to shade in Arabidopsis: role of hormone, clock, and light signaling. Mol Plant 5, 619-628.
Sidler, W., 1994. Phycobilisome and phycobiliprotein structures, in: Bryant, D. (Ed.), The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 139-216.
Singh, S.P., Montgomery, B.L., 2012. Reactive oxygen species are involved in the morphology-determining mechanism of Fremyella diplosiphon cells during complementary chromatic adaptation. Microbiol 158, 2235-2245.
Sobczyk, A., Schyns, G., Tandeau de Marsac, N., Houmard, J., 1993. Transduction of the light signal during complementary chromatic adaptation in the cyanobacterium Calothrix sp. PCC 7601: DNA-binding proteins and modulation by phosphorylation. EMBO J 12, 997-1004.
Song, J.Y., Cho, H.S., Cho, J.I., Jeon, J.S., Lagarias, J.C., Park, Y.I., 2011. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 108, 10780-10785.
Stomp, M., Huisman, j., Jongh, F., Veraart, A.J., Gerla, D., Rijkeboer, M., Ibelings, B.W., Wollenzien, U.I.A., Stal, L.J., 2004. Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature, 104–107.
Stomp, M., van Dijk, M.A., van Overzee, H.M., Wortel, M.T., Sigon, C.A., Egas, M., Hoogveld, H., Gons, H.J., Huisman, J., 2008. The timescale of phenotypic plasticity and its impact on competition in fluctuating environments. American Naturalist 172, 169-185.
Strauss, H.M., Hughes, J., Schmieder, P., 2005. Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochem 44, 8244-8250.
Tandeau de Marsac, N., 1977. Occurrence and nature of chromatic adaptation in cyanobacteria. J Bacteriol 130, 82-91.
Tandeau de Marsac, N., 1983. Phycobilisomes and complementary chromatic adaptation in cyanobacteria. Bull Inst Pasteur 81, 201-254.
Tandeau de Marsac, N., Houmard, J., 1993. Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol Rev 104, 119–190.
Tandeau de Marsac, N., Mazel, D., Damerval, T., Guglielmi, G., Capuano, V., Houmard, J., 1988. Photoregulation of gene expression in the filamentous cyanobacterium Calothrix sp. PCC 7601: light harvesting complexes and cell differentiation. Photosyn Res 18, 99-132.
Tandeau de Marsac, N.T., Castets, A.M., Cohen-Bazire, G., 1980. Wavelength modulation of phycoerythrin synthesis in Synechocystis sp. 6701. J Bacteriol 142, 310-314.
Terauchi, K., Montgomery, B.L., Grossman, A.R., Lagarias, J.C., Kehoe, D.M., 2004. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol Microbiol 51, 567-577.
Ulijasz, A.T., Cornilescu, G., von Stetten, D., Cornilescu, C., Velazquez Escobar, F., Zhang, J., Stankey, R.J., Rivera, M., Hildebrandt, P., Vierstra, R.D., 2009. Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J Biol Chem 284, 29757-29772.
Ulijasz, A.T., Vierstra, R.D., 2011. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol 14, 498-506.
van Waasbergen, L.G., Dolganov, N., Grossman, A.R., 2002. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J Bacteriol 184, 2481-2490.
Vierstra, R.D., Zhang, J. 2011. Phytochrome signaling: solving the Gordian knot with phytochrome relatives. Trends Plant Sci 16, 417-426.
Vogelmann, T.C., Scheibe, J., 1978. Action spectrum for chromatic adaptation in the blue-green alga Fremyella diplosiphon. Planta 3, 233-239.
Wagner, J.R., Brunzelle, J.S., Forest, K.T., Vierstra, R.D., 2005. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325-331.
Wenden, B., Kozma-Bognar, L., Edwards, K.D., Hall, A.J., Locke, J.C., Millar, A.J., 2011. Light inputs shape the Arabidopsis circadian system. Plant J 66, 480-491.
Wilde, A., Churin, Y., Schubert, H., Borner, T., 1997. Disruption of a Synechocystis sp. PCC 6803 gene with partial similarity to phytochrome genes alters growth under changing light qualities. FEBS Lett 406, 89-92.
Wilde, A., Fiedler, B., Borner, T., 2002. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol Microbiol 44, 981-988.
Wu, S.H., Lagarias, J.C., 2000. Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochem 39, 13487-13495.
Yeh, K.C., Lagarias, J.C., 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95, 13976-13981.
Yeh, K.C., Wu, S.H., Murphy, J.T., Lagarias, J.C., 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505-1508.
Yoshihara, S., Katayama, M., Geng, X., Ikeuchi, M., 2004. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant & Cell Physiol 45, 1729-1737.
Yoshihara, S., Shimada, T., Matsuoka, D., Zikihara, K., Kohchi, T., Tokutomi, S., 2006. Reconstitution of blue-green reversible photoconversion of a cyanobacterial photoreceptor, PixJ1, in phycocyanobilin-producing Escherichia coli. Biochem 45, 3775-3784.
Yoshihara, S., Suzuki, F., Fujita, H., Geng, X.X., Ikeuchi, M., 2000. Novel putative photoreceptor and regulatory genes Required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant & Cell Physiol 41, 1299-1304.
11/20/12