GREEN FLUORESCENT PROTEIN:
A MOLECULAR MICROSCOPE
Marc Zimmer
Department of Chemistry
Connecticut College
New London, CT 06320
mzim@conncoll.edu
Introduction
In 1674 Anton von Leeuwenhoek invented the simple microscope, and it opened up a whole new world. Suddenly one could see things that had never been visible before. His microscope was able to magnify objects up to 300 times, and he was one of the first people to observe spermatozoa, blood cells, bacteria, and muscle fibers.
Green Fluorescent Protein (GFP) and GFP-like proteins have become the microscope of the twenty-first century. Every month more than 200 papers are published reporting yet another way GFP has been put to work. In most cases, GFP can be used in a way very similar to a microscope; it can show us when a protein is made, and what its movements are.
In this module I will introduce the biology, chemistry and uses of GFP and GFP-like proteins. In honor of the 2008 Nobel Prize in Chemistry, the whole October 2009 issue of Chemical Society Reviews (Vol. 38, pp. 2813-2963) is devoted to GFP. This is an excellent resource for anyone wanting more detailed information about GFP than is presented in this module.
My books, Illuminating Disease: An Introduction to Green Fluorescent Protein (Zimmer, 2015a), and Bioluminescence: Nature and Science at Work (Zimmer, 2015b) are also good general references.
Aequorea Bioluminescence
The crystal jellyfish (Aequorea victoria) has about three hundred photoctyes located on the edge of its umbrella, when stimulated they give off green light (Figure 1).
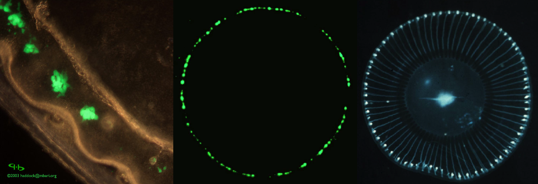
Figure 1. Aequorea victoria photocytes are located on the edge of the umbrella. The left image shows a microscopic view of some photocytes. The central photograph shows the bioluminescence of the photocytes, while the right hand image shows the jellyfish under visible light. The blue in the photograph on the right is not due to bioluminescence or fluorescence, it is due to visible light reflection. [Left Image: from The Bioluminescence Web Page (Haddock and Case). Right and middle images: courtesy of Osamu Shimomura]
The jellyfish does not use the common luciferase/luciferin system to produce its bioluminescence. Instead two proteins are involved in Aequorea bioluminescence; a calcium binding protein (aequorin) and a green fluorescent protein (Morise et al., 1974). In the absence of GFP, aequorin gives off blue light upon binding calcium; however, in the jellyfish, radiationless energy transfer occurs.
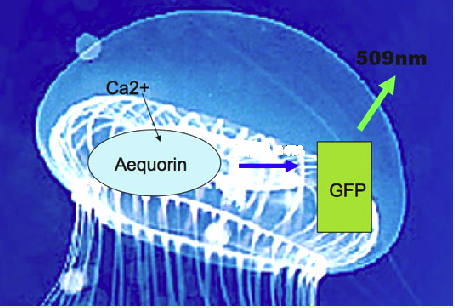
Figure 2. Upon binding calcium, aequorin generates an electronically excited product that undergoes radiationless energy transfer (blue arrow) to the GFP fluorescent state, which emits the green light (509 nm) (see Figure 1).
Green Fluorescent Protein Structure
The amino acid sequence of GFP was first reported in 1992 (Prasher et al., 1992). It has 238 amino acids (Figure 3).

Figure 3. Nucleotide and amino acid sequence of GFP cDNA. The chromophore forming amino acids are highlighted in green.
The GFP chromophore is formed in an autocatalytic cyclization of the tripeptide 65SYG67 sequence, it does not require a cofactor, and is followed by an oxidation of the intrinsically formed structure. This means that GFP fluoresces in the absence of any other proteins, substrates, or cofactors (Scheme 1).
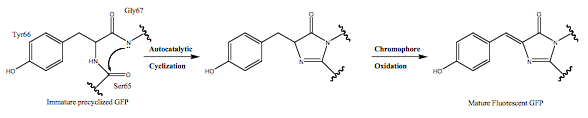
Scheme 1. Autocatalytic chromophore formation observed in fluorescent proteins.
GFPs have a unique 11 ß-sheet barrel-like structure with a diameter of about 24Å and a height of 42Å (Figure 4). The ß-sheets form the walls of the barrel and an alpha helix runs diagonally through it. The chromophore is in the center of the 11 ß-sheets and is linked by the
 -helical stretch that runs through the center of the barrel. The Protein Data Bank currently lists over 200 crystal structures of GFP, its mutants and GFP-like proteins. Even though many of the mutants and GFP-like proteins have very different spectral properties, their gross structural features are remarkably similar to wild-type GFP.
-helical stretch that runs through the center of the barrel. The Protein Data Bank currently lists over 200 crystal structures of GFP, its mutants and GFP-like proteins. Even though many of the mutants and GFP-like proteins have very different spectral properties, their gross structural features are remarkably similar to wild-type GFP.
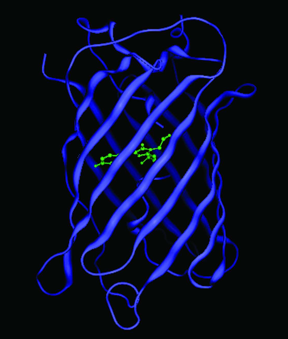
Figure 4. Crystal structure of GFP. The chromophore is shown in green, and is located in the center of the ß-barrel. Coordinates obtained from the Protein Data Bank (1GFL).
The Fluorescent Protein Palette
By mutating Tyr66, the central chromophore forming residue, to His or Trp, blue and cyan fluorescent mutants can be obtained. Many other colors have been accessed by mutating the chromophore forming residues, and the residues surrounding the chromophore. However, the color palette obtained by mutating GFP was insufficient, and researchers went back to nature to find more fluorescent proteins.
Once it was discovered that aequorin and GFP had evolved separately, and that fluorescent proteins did not necessarily have to be associated with other chemiluminescent proteins, over 150 distinct fluorescent or colored GFP-like proteins have been reported. GFP-like fluorescent proteins (FPs) have been found in marine organisms ranging from chordates (e.g., amphioxus) to cnidarians (e.g., corals and sea pansies). No FPs have been found in terrestrial organisms. The majority of GFP containing organisms are non-bioluminescent. These fluorescent proteins can be divided into seven groups according to their color and chromophore structure (Labas et al., 2002). [see Figures 5 and 6]
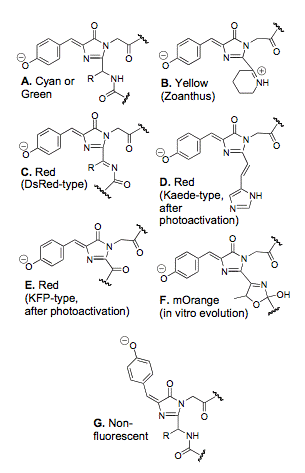
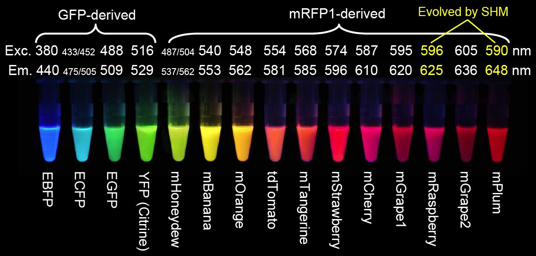
Figure 6. Fluorescent proteins discussed above, and the mFruit FPs derived by Roger Tsien from mRFP1 (Red Fluorescent Protein) (Shaner et al., 2004), and by somatic hypermutation (SHM) (Wang et al., 2004). E stands for enhanced versions of GFP, m are monomeric proteins, and tdTomato is a head-to-tail dimer (image from Roger Y. Tsien Nobel Lecture 2008.)
Fluorescent Protein Uses
In the last fifteen years green fluorescent protein (GFP) has changed from a nearly unknown protein to a commonly used molecular imaging tool in biology, chemistry, genetics and medicine. Probably the best indicator of the utility of GFP and GFP-like proteins is the 2008 chemistry Nobel Prize that was awarded to Profs. Shimomura, Chalfie and Tsien for "the discovery and development of the green fluorescent protein", and the fact that in 2004 about 50%, 35%, 60% and 20% of the articles in the journals Cell, Development, Journal of Cell Biology, and Neuron mentioned or used GFP-like proteins (Chalfie & Kain, 2006).
GFPs and GFP-like proteins (i.e., chromoproteins and fluorescent proteins) are particularly useful due to their stability, and the fact that the chromophore is formed in an autocatalytic cyclization of the 65SYG67 sequence, which does not require a cofactor. This means that unlike most other bioluminescent reporters, GFP fluoresces in the absence of any other proteins, substrates, or cofactors. Thus, GFP can be used as a genetic tracer molecule (Figure 7). Furthermore, it appears that fusion of GFP to a protein does not alter the function or location of the protein.
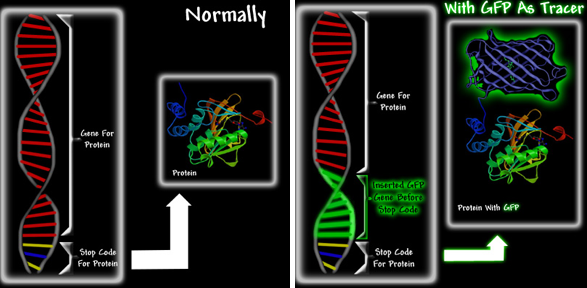
Figure 7. A schematic showing how GFP can be used as a fluorescent tag. Proteins are extremely small and cannot be seen, even under an electron microscope. However, if one could somehow link GFP to a specific protein, one would be able to see the green fluorescence of the GFP that is attached to the protein. It would be a bit like attaching a light bulb to the protein. Left: When new proteins are required, protein production is activated. The gene is read, and the protein is manufactured. At the end of the gene is a message called a stop codon, which ends protein production. The manufacture of proteins using the instructions from the gene is called protein expression. Right: The GFP gene is inserted at the end of the protein gene, but before the stop codon. The resultant GFP-fusion protein shown in the right image is fluorescent, and can functionally replace the original protein. Upon expression the GFP-chimera will be green fluorescent, thereby allowing researchers to track both its expression and movement.
A literature search for papers with "green fluorescent protein" in the title, abstract or keywords found one paper published in 1990, 1441 in 1999, and 4585 in 2009. Two books for the non-scientist have been written about GFP, fluorescent proteins have appeared in numerous art exhibits, and a Google search reveals more than 150,000 GFP images. In the next two paragraphs I will briefly describe two uses. Hopefully these two examples demonstrate some of the utility, beauty and versatility of fluorescent protein-based-techniques.
Cell growth occurs through an ordered sequence of events, i.e., the cell cycle, which consists of four distinct phases, G1, S, G2 and M phase. In February 2008, Atsushi Miyawaki from the RIKEN Basic Science Institute reported that his group has developed a method, Fucci (fluorescent, ubiquitination-based cell cycle indicator), which will allow cell cycle researchers to visualize cell cycle progression (Sakaue-Sawano et al., 2008). Fucci modified cells are yellow at the start of replication, switch to green during S phase, and to red during G1 (Figure 8).
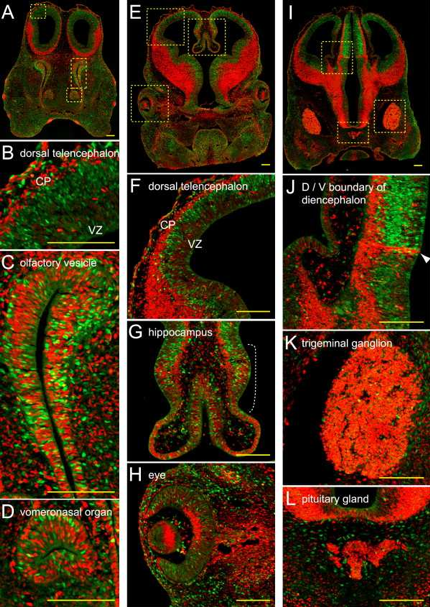
Figure 8. To demonstrate the utility of Fucci, researchers created a Fucci mouse. The colored images shown here demonstrate the equilibrium between cell differentiation and cell proliferation that occurs during the development of the mouse head. The red and green signals appear to be well balanced at the embryonic stage, but the overall ratio of green-to-red signal decreases as the mice grow (Sakaue-Sawano et al., 2008).
When human T-cells bump into each other they form a sticky strand that connects the two cells. These strands dubbed "membrane nanotubes" by the Imperial College scientists who discovered them can connect two T-cells that are several cell lengths apart (Sowinski et al., 2008). By infecting a T-cell with HIV containing GFP labelled proteins the researchers were able to show that HIV proteins travel down the nanotubes from infected to non-infected cells (Figure 9). These nanotubes maybe part of the reason HIV is so effective at spreading rapidly within host bodies.
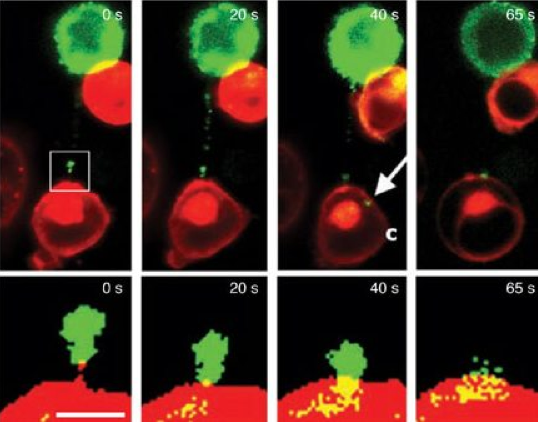
Figure 9. Time-lapse imaging of GFP tagged proteins moving along a membrane nanotube connecting infected with uninfected T cells. The boxed region in the top left image is enlarged in the images in the bottom panel to show that the protein-GFP reaches the initially uninfected T cell (Sowinski et al., 2008). The arrow labeled C shows the GFP chimera in the cytoplasm of an initially uninfected cell.
New Techniques Based on GFP
Optogenetics: An On/Off Switch for Individual Neurons
Until recently, neuroscientists would stimulate brain cells with electrodes, but even with the finest electrodes they could never activate single neurons. Now, thanks to optogenetics, scientists can use light and an algae protein to turn individual neurons on and off instantly. Some of the earliest work in optogenetics was done by Karl Deisseroth and Ed Boyden while they were working in Roger Tsien's lab (UC San Diego). So, it is not surprising that optogenetics can be viewed as an outgrowth of GFP technology, and that fluorescent proteins are routinely used to show which neurons have been modified to become neural on/off switches.
In algae, the function of the protein channelrhodopsin is to allow calcium ions to enter the algae cells when they are exposed to blue light. In 2005, Karl Deisseroth, a professor in the departments of bioengineering and psychiatry at Stanford University, took the channelrhodopsin gene and inserted it into a single neuron of Caenorhabditis elegans, a worm often used in scientific studies. To the same worm, he added the gene for halorhodopsin, a protein found in very primitive bacteria that thrive in salt flats, where the protein allows the chloride ion to enter the bacterium upon exposure to yellow light. Negative chloride and positive calcium ions combine to neutralize each other. The neuron in Deisseroth's worm now had an on switch (blue light -> calcium ions) and an off switch (yellow light -> chloride ions). C. elegans has 302 neurons, and their functions are well known, which allowed Deisseroth to modify a neuron responsible for movement. With blue light the worm wiggled, and with yellow light it stopped (Boyden et al., 2005). In 2010, Nature Methods announced that Optogenetics was its method of the year.
Spinach: An RNA Mimic of GFP
Numerous studies of green fluorescent protein's photochemistry have utilized model compounds synthesized to mimic the proteins chromophore. The most commonly used is 4-hydroxybenzlidene imidazolinone (HBI) (Figure 10).
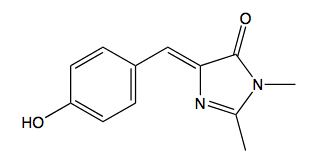
Figure 10. 4-hydroxybenzlidene imidazolinone (HBI)
These organic model compounds only fluoresce when their dihedral freedom is severely restricted. Page et al. (2011) have designed two new ligands based on HBI, DMHBI and DMFBI (Figure 11).
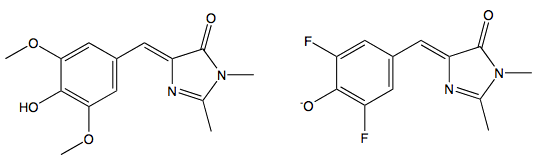
Figure 11. Left: 3,5-dimethoxy-4-hydroxybenzylidene imidazolinone (DMHBI). Right: 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI)
These fluorophores selectively bind specific RNA sequences that were found by ten rounds of systematic evolution of ligands by exponential enrichment (SELEX). When bound to the RNA aptamer, their dihedral freedom is sufficiently restricted so that they fluoresce. Like GFP, DMHBI is predominantly found in the phenolic form, the brighter DMFBI mimics enhanced GFP (EGFP) (Heim et al., 1995), and is anionic under standard conditions. The DMFBI-RNA complex has been called Spinach, it is the first fluorescent RNA tag that is both selective and non-toxic. It only fluoresces when bound to RNA, is resistant to photobleaching, and has been used to follow RNA tagged molecules as they move through cells (Page et al., 2011).
Brainbow
Jeff Lichtman and Joshua Sanes, researchers at the Harvard Brain Center, have created transgenic mice with fluorescent multicolored neurons. The photographs of their mouse brains that appeared in the Nov. 1 (2007) issue of Nature could be displayed in the Museum of Modern Art.
In the Brainbow mice, the Harvard researchers have introduced genetic machinery that randomly mixes green, cyan and yellow fluorescent proteins in individual neurons, thereby creating a palette of 90 distinctive hues and colors. “The technique drives the cell to switch on fluorescent protein genes in neurons more or less at random,” says Jean Livet, the postdoctoral researcher responsible for most of the laboratory work that resulted in the brainbow mice. “You can think of Brainbow almost like a slot machine in its generation of random outcomes.”
Using a Brainbow of colors, researchers can now map the neural circuits of the brain (Weissman and Pan, 2015).
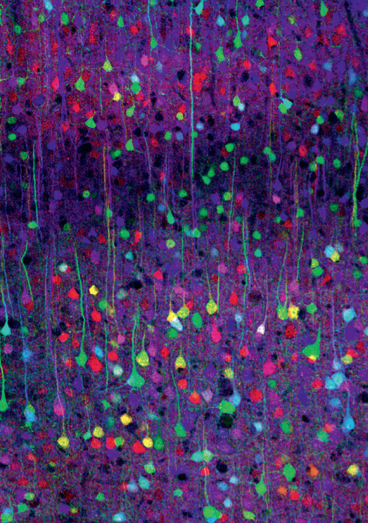
Figure 12. Mouse cerebral cortex imaged with Brainbow (Confocal image by Tamily Weissman. Mouse by Jean Livet and Ryan Draft.)
In a 2011 profile of Josh Sanes, The Scientist asked him how techniques like Brainbow are useful. “They’re really good for getting pictures on the covers of journals and books,” he joked before describing how his techniques allows the average neuroscientist to do what Cajal did more than a century ago. “Seriously, they allow you to distinguish individual neurons out of a morass. Just like Cajal did with his Golgi stain. If you stain a single neuron in its entirety you can learn about its shape and its connections. If you label 100 neurons in a single mouse, you learn much more than if you study 100 mice with one neuron labeled in each. Cajal was an incredible genius in that he could look at one neuron in each of 100 mice, and then go home and draw a picture that synthesized all of that information. And almost always he got that right. Anyone else would have gotten it wrong, because you can’t register all those variations against each other unless you are supernaturally brilliant like Cajal.” (Hopkin, 2011)
Conclusion
GFP is used in thousands of experiments every day, a large number of them don't work, and most never make it into the scientific literature. Very few are ever reported in the popular press; only the ones that make good copy are. They are rarely the most important ones. Nonetheless, they illustrate that GFP is becoming a commonplace scientific tool, just like a microscope, an electrophoresis gel, or even a test tube.
References
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., and Deisseroth, K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience 8, 1263-1268.
Chalfie, M., & Kain, S. R. (2006). Green fluorescent protein: Properties, applications and protocols. Second ed. Hoboken, New Jersey: John Wiley & Sons.
Haddock, S.H.D. & Case, J.F., The Bioluminescence Web Page.
Heim, R., Cubitt, A. B., and Tsien, R.Y. (1995), Improved green fluorescence. Nature 373, 663-664.
Hopkin, K. (2011) Critical Connections, Scientist 25, 54-56.
Labas, Y. A., Gurskaya, N. G., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Lukyanov, S. A., and Matz, M. V. (2002). Diversity and evolution of the green fluorescent protein family. Proceedings of the National Academy of Sciences 99, 4256-4261.
Morise, H., Shimomura, O., Johnson, F. H., & Winant, J. (1974). Intermolecular energy transfer in bioluminescent systems of Aequorea. Biochemistry 13, 2656-2662.
Paige, J. S., Wu, K. Y., and Jaffrey, S. R. (2011). RNA Mimics of Green Fluorescent Protein. Science 333, 642-646.
Prasher, D. C., Eckenrode, V. K., Aequorea victorea green fluorescent protein. Gene 111, 229-233.
Sakaue-Sawano, A., Kurokawa, H., Morimura, T., Hanyu, A., Hama, H., Osawa, H., Kashiwagi, S., Fukami, K., Miyata, T., Miyoshi, H., Imamura, T., Ogawa, M., Masai, H., and Miyawaki, A. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487-498.
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N. G., Palmer, A. E., & Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology 22, 1567-1572.
Sowinski, S., Jolly, C., Berninghausen, O., Purbhoo, M. A., Chauveau, A., Kohler, K., Oddos, S., Eissmann, P., Brodsky, F. M., Hopkins, C., Onfelt, B., Sattentau, Q., and Davis, D. M. (2008). Membrane nanotubes physically connect t cells over long distances presenting a novel route for hiv-1 transmission. Nature Cell Biology 10, 211-219.
Wang, L., Jackson, W. C., Steinbach, P. A., & Tsien, R. Y. (2004). Evolution of new nonantibody proteins via iterative somatic hypermutation. Proceedings of the National Academy of Sciences 101, 16745-16749.
Weissman, T. A., and Pan, Y. A. (2015) Brainbow: New Resources and Emerging Biological Applications for Multicolor Genetic Labeling and Analysis, Genetics 199, 293-306.
Zimmer, M. (2015a) Illuminating disease : An introduction to green fluorescent proteins, Oxford University.
Zimmer, M (2015b) Bioluminescence: Nature and Science at Work, Twenty-First Century Books.
04/21/10
08/24/11
01/25/16