RECOMBINATIONAL DNA REPAIR
Kendric C. Smith
Emeritus Professor, Radiation Oncology (Radiation Biology)
Stanford University School of Medicine
800 Blossom Hill Road, Unit R169, Los Gatos, CA 95032
kendric@stanford.edu
web.stanford.edu/~kendric/
The two major systems for the repair of ultraviolet (UV) radiation-induced DNA damage in cells are nucleotide excision repair (Petit and Sancar, 1999), and recombinational DNA repair (Kuzminov, 1999; Smith, 2004). In nucleotide excision repair, damaged nucleotides (i.e., pyrimidine dimers) are recognized and cut out, and the resulting hole is patched by DNA polymerase l, using the DNA strand opposite the gap as the template. The patch is then joined to the main strand by DNA ligase. This process is very accurate, and does not produce mutations. The sheer volume of publications on "cut and patch" nucleotide excision repair (e.g., Petit and Sancar, 1999) has generated the impression that cells possess only this simple repair system.
Quite to the contrary, recombinational DNA repair is critical for the survival of UV radiation-damaged cells. It accounts for about 50% of the survival of UV irradiated Escherichia coli (see below). It is a very complicated process that requires two DNA duplexes, and the exchange of a strand of DNA from one DNA duplex to the other (Rupp et al., 1971; Howard-Flanders and Rupp, 1972; Ganesan, 1974; Wang and Smith, 1984), and it produces mutations.
The first indication that nucleotide excision repair ("cut and patch") is NOT the only mechanism by which cells repair damage to their DNA, was the observation that bacterial cells deficient in nucleotide excision repair (i.e., uvrA) or in genetic recombination (i.e., recA) are very sensitive to UV radiation, and show a similar level of survival after UV irradiation. A double mutant (uvrA recA), however, is much more sensitive to UV irradiation than either of the single mutants (Figure 1). From the most fundamental principles of radiation biology and genetics, these data argue that, (a) these two systems, i.e., coded by the uvrA and the recA genes, function largely independently of each other, and (b) they are of about equal importance to the survival of UV-irradiated cells of E. coli K-12. These studies led to the discovery of postreplication repair (see below).
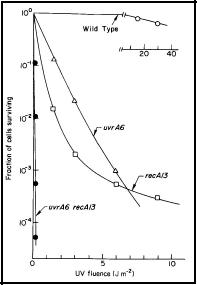
Figure 1. UV radiation survival curves for DNA repair deficient mutants of E. coli K-12. The uvrA6 mutation blocks nucleotide excision repair, and the recA13 mutation blocks recombinational DNA repair. Note that the double mutant, uvrA6 recA13 is very much more sensitive to UV radiation than either of the single mutants, indicating that the two single mutants are involved in separate pathways of DNA repair. Since the two single mutants have about the same sensitivity, it indicates that nucleotide excision repair and recombinational DNA repair are of about equal importance to the survival of UV irradiated E. coli. [Modified from Howard-Flanders and Boyce, 1966]
Although ignored in most reviews on DNA repair, there is a pathway of nucleotide excision repair that is dependent upon recombination (see below). This occurs when lesions are produced in the portion of the chromosome that was replicated PRIOR to UV irradiation, and therefore, two DNA duplexes are present in the region of the lesion (Figure 2).
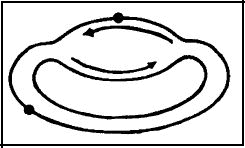
Figure 2. Schematic of DNA replication in E. coli with lesions (large dots), both in the DNA that was replicated prior to UV irradiation, where two DNA duplexes exist, and in that portion of the chromosome prior to replication, where only one DNA duplex exists. The problems and opportunity for recombinational DNA repair in these two regions of the chromosome are markedly different.
A third type of recombinational DNA repair is the repair of DNA double-strand breaks that are produced metabolically after UV irradiation (see module on DNA Double-Strand Breaks).
Postreplication Repair
The DNA synthesized immediately after UV irradiation in excision repair-deficient cells (and also wild-type cells; see below) of E. coli K-12 has discontinuities when assayed in alkaline sucrose gradients. The mean length of newly synthesized DNA approximates the distance between pyrimidine dimers in the parental strand. With further incubation of the cells, however, these discontinuities disappear, and the DNA approximates the molecular size of that from unirradiated control cells (Rupp and Howard-Flanders, 1966; Howard-Flanders et al., 1968). The exchanges envisioned by this type of repair resemble those involved in genetic recombination (Rupp et al., 1971; Rupp and Howard-Flanders, 1968). This prediction has been verified by demonstrating that recA cells are deficient in the production of normal length DNA from the small pieces of DNA synthesized immediately after UV irradiation (Smith and Meun, 1970; Sedgwick, 1975).
When DNA synthesis proceeds along a damaged template, synthesis halts at the site of a non-coding lesion, and then resumes downstream from the lesion (i.e., at the next DnaG primase-binding site), leaving gaps in the newly synthesized daughter strand opposite the UV radiation-induced lesion in the parental strand (Rupp and Howard-Flanders, 1968). The fact that photoreactivation after UV irradiation in a uvrA strain stimulated gap filling, is taken as further evidence that a large proportion of the DNA daughter-strand gaps are opposite pyrimidine dimers (Bridges and Sedgwick, 1974).
[see Photoreactivation module]
The dimers that are opposite DNA daughter-strand gaps are no longer subject to excision, since this process requires an intact complementary strand (Jansz, Pouwels and Van Rotterdam, 1963; Yarus and Sinsheimer, 1984). Only after the gaps are filled by sister-strand exchanges will the dimers again be subject to excision repair.
These gaps in the daughter strands, which average 1000 nucleotides in length (Iyer and Rupp, 1971), are subsequently repaired in recombination proficient strains by transferring the appropriate sections of DNA from the parental strands into the daughter strands. This transfer of parental strands into daughter strands has been confirmed by direct measurement (Rupp et al., 1971; Howard-Flanders and Rupp, 1972; Ganesan, 1974; Wang and Smith, 1984). Although most studies on postreplication repair have been performed in excision repair deficient cells, this type of repair is fully operative in wild-type cells (Smith and Meun, 1970; Rupp, Iyer and Zipser, 1973; Howard-Flanders and Rupp, 1981).
Although postreplication repair (i.e., the repair of DNA daughter-strand gaps) is completely dependent upon the recA gene, mutations in the recB and recC genes do NOT cause a deficiency in the repair of DNA daughter-strand gaps (Smith and Meun, 1970). However, the recB gene is known to function in the repair of DNA double-strand breaks that are formed metabolically after UV irradiation in E. coli (Wang and Smith, 1975). In fact, unrepaired DNA double-strand breaks appear to be the major cause of lethality in UV-irradiated wild-type bacteria (Bonura and Smith, 1975a,b). The repair of metabolically-produced DNA double-strand breaks constitutes a second type of recombination repair that is distinct from the repair of DNA daughter-strand gaps, i.e., it is recBC-dependent (Wang and Smith, 1975, 1986).
[see module on
DNA Double-Strand Breaks]
Multiple Pathways of Postreplication Repair
Three pathways are known for the repair of DNA daughter-strand gaps, i.e., the recF-dependent, the recF-independent, and the umuCD-dependent pathways. Much of postreplication repair is constitutive (Ganesan and Smith, 1972; Sedgwick, 1975), but a portion (i.e., umuCD) is inducible by UV radiation, and is responsible for UV radiation mutagenesis (see below). Each of these pathways is recBC-independent (Smith and Meun, 1970).
RecF Pathway: About half of the DNA daughter-strand gaps are repaired by a recF-dependent process (Wang and Smith, 1975; Ganesan and Seawell, 1975; Kato, 1977; Tseng, Hung and Wang, 1994). The involvement of the recF gene suggests that the recF pathway of homologous recombination may be involved in this repair process. The RecF protein is one of at least three single-strand DNA binding proteins, along with the RecA and Ssb proteins (Madiraju and Clark, 1991).
The repair of daughter-strand gaps by the recF-dependent and the recF-independent process (see below) is accompanied by the transfer of DNA lesions from the parental strand to the daughter strand (Ganesan, 1974; Wang and Smith, 1984). This occurs about 50% of the time in E. coli (Ganesan, 1974), and appears to be due to the random resolution of the Holliday junction (e.g., Sigal and Alberts, 1972), an intermediate in recombination.
RecF-Independent Pathway: The fact that a uvrB recF stain is not as deficient in the repair of daughter-strand gaps as is a uvrB recA strain suggested that a second pathway must exist for the repair of daughter-strand gaps (Wang and Smith, 1975). This conclusion was supported by studies using an insertion mutation of recF (recF332::Tn3) to ensure that the earlier results were not due to leakiness in the original recF143 mutation. The recF-independent pathway is also independent of the recBC genes, and is constitutive (Sharma and Smith, 1985). Studies using 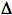 polA mutants, indicate that the polA gene (DNA polymerase l) plays a major role in the recF-independent repair of daughter-strand gaps. Studies on different polA mutants (i.e., polA1, polAex2,
polA mutants, indicate that the polA gene (DNA polymerase l) plays a major role in the recF-independent repair of daughter-strand gaps. Studies on different polA mutants (i.e., polA1, polAex2, 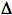 polA, etc.) suggest that it is the 5'
polA, etc.) suggest that it is the 5'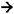 3' exonuclease activity of DNA polymerase l that plays a major role in the repair of daughter-strand gaps (Sharma and Smith, 1987).
3' exonuclease activity of DNA polymerase l that plays a major role in the repair of daughter-strand gaps (Sharma and Smith, 1987).
Furthermore, since DNA polymerase is known to be involved in the joining of Okazaki fragments synthesized in the lagging strand of unirradiated cells, this raises the possibility that the daughter-strand gaps formed in the lagging strand of UV irradiated cells may be selectively repaired by the recF-independent, polA-dependent pathway, while the daughter-strand gaps formed in the leading strand (i.e., presumably longer gaps) may be repaired by the recF-dependent pathway (Liu, Cheng and Wang, 1998).
UmuC Pathway: Since a uvrA 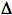 polA recF strain is not quite as deficient in the repair of daughter-strand gaps as is a uvrA recA strain (Sharma and Smith, 1987), it suggests that a third pathway must exist for the repair of daughter-strand gaps. Consistent with this observation, a small fraction of the repair of daughter-strand gaps is dependent upon the umuC gene, but is independent of the recF and recBC genes (Wang and Smith, 1985). A uvrA
polA recF strain is not quite as deficient in the repair of daughter-strand gaps as is a uvrA recA strain (Sharma and Smith, 1987), it suggests that a third pathway must exist for the repair of daughter-strand gaps. Consistent with this observation, a small fraction of the repair of daughter-strand gaps is dependent upon the umuC gene, but is independent of the recF and recBC genes (Wang and Smith, 1985). A uvrA 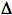 polA recF umuC strain has not yet been tested to see if it as deficient as a uvrA recA strain in the repair of daughter-strand gaps.
polA recF umuC strain has not yet been tested to see if it as deficient as a uvrA recA strain in the repair of daughter-strand gaps.
The UmuC and UmuD proteins combine, after the selective cleavage of the UmuD protein by RecA, to form an error-prone polymerase (UmuD'2UmuC), polV (Tang et al., 1999; Ferentz, Walker and Wagner, 2001), which can synthesize past lesions in DNA. This is consistent with the fact that umuC controls all of UV radiation mutagenesis (Kato and Shinoura, 1977). A umuC mutation, however, has only a partial effect on spontaneous mutagenesis (Sargentini and Smith, 1981), and on X-ray mutagenesis (Sargentini and Smith, 1989). [see module on
UV Radiation and Spontaneous Mutagenesis]
Nucleotide Excision Repair
There are two pathways of nucleotide excision repair. One pathway is DNA polymerase l dependent, growth medium independent (i.e., macromolecular synthesis is not required), and it produces short repair patches (about 20 nucleotides long). This pathway requires only one DNA duplex (Petit and Sancar, 1999).
The second excision repair process, long-patch excision repair, which requires two DNA duplexes, is largely ignored by reviewers (e.g., Hanawalt, 2001). Nevertheless, this excision repair pathway does exist, and it has been confirmed by other authors (e.g., Youngs et al., 1974). It is dependent upon the recA gene, it is growth medium dependent (i.e., macromolecular synthesis is required), and it produces long repair patches (1500-9000 nucleotides long) (Cooper and Hanawalt, 1972a, b; Cooper, 1982). Long-patch excision repair also requires the recF gene (Hanawalt et al., 1982), but does NOT require the recBC genes (Hanawalt, Cooper and Smith, 1981).
When wild-type cells are allowed to repair their DNA after UV irradiation in the presence of chloramphenicol to inhibit the synthesis of induced proteins, only about 80% of the dimers are excised (Lin, Kovalsky and Grossman, 1997). Similarly, a recA mutant, which is deficient in the induction of proteins after UV irradiation, only excises about 80% of the dimers compared to a wild-type strain (Shlaes, Anderson, and Barbour, 1972). The early repair seems to be short-patch excision repair, which occurs immediately after UV irradiation, and is controlled by DNA polymerase l (Cooper and Hanawalt, 1972a), while the induced repair appears to be the long-patch system that is controlled by recA (Cooper, 1982). Additional copies of the UvrA protein (Kenyon and Walker, 1981) and the UvrB protein (Schendel, Fogliano, and Strausbaugh, 1982) are synthesized after UV irradiation, and may be relevant to the inducible long-patch excision repair process.
The excision repair that occurs in cells that contain completely replicated chromosomes, i.e., where only one DNA duplex is present per chromosome, is not dependent upon recA. In this situation, classical nucleotide excision repair occurs, i.e., without strand exchanges. The excision repair that functions in the part of the chromosome that was replicated before UV irradiation (i.e., where two DNA duplexes exist, Figure 1), is recA-dependent (Smith and Sharma, 1987).
The similarities between the genetic requirements for long-patch excision repair and the repair of DNA daughter-strand gaps, i.e., the requirement for recA and recF, but not recBC, and the requirement for sister DNA duplexes, suggests that the mechanisms for these two repair processes are similar, i.e., requiring strand exchanges. The only significant difference between these two processes is the manner in which the gaps in the sister duplexes are formed, i.e., by excision or by replication bypass (Smith and Sharma, 1987).
For a discussion of excision repair in mammalian cells, see the module Nucleotide Excision Repair in Human Cells.
Summary and Conclusions
It is unfortunate that the importance of recombinational DNA repair is being ignored in many articles on DNA repair. Furthermore, most reviewers make no distinction between the repair events that take place in the two different parts of the chromosome, i.e., the part of the chromosome that was replicated BEFORE UV irradiation, where two DNA duplexes exist, and the part of the chromosome that contains only ONE DNA duplex, and is replicated after UV irradiation. Clearly the problems and the opportunities for DNA repair are quite different in these two regions of the chromosome.
References
Bonura T, Smith KC. 1975a. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J Bacteriol 121:511-517.
Bonura T, Smith KC. 1975b. Quantitative evidence for enzymatically-induced DNA double-strand breaks as lethal lesions in UV-irradiated pol+ and polA1 strains of E. coli K-12. Photochem Photobiol 22:243-248.
Bridges BA, Sedgwick SG. 1974. Effect of photoreactivation on the filling of gaps in deoxyribonucleic acid synthesized after exposure of Escherichia coli to ultraviolet light. J Bacteriol 117:1077-1081.
Cooper PK. 1982. Characterization of long patch excision repair of DNA in ultraviolet-irradiated Escherichia coli: an inducible function under Rec-Lex control. Mol Gen Genet 185:189-197.
Cooper PK, Hanawalt PC. 1972a. Role of DNA polymerase l and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci USA 69:1156-1160.
Cooper PK, Hanawalt PC. 1972b. Heterogeneity of patch size in repair replicated DNA in Escherichia coli. J Mol Biol 67:1-10.
Ferentz AE, Walker GC, Wagner G. 2001. Converting a DNA damage checkpoint effector (UmuD'2C) into a lesion bypass polymerase (UmuD'2C). The EMBO J
20:4287-4298.
Ganesan AK. 1974. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K-12. J Mol Biol 87:103-119.
Ganesan AK, Seawell PC. 1975. The effect of lexA and recF mutations on post-replication repair and DNA synthesis in Escherichia coli K-12. Mol Gen Genet 141:189-205.
Ganesan A K, Smith KC. 1972. Requirement for protein synthesis in rec-dependent repair of deoxyribonucleic acid in Escherichia coli after ultraviolet or X irradiation. J Bacteriol 111:575-585.
Hanawalt PC. 2001. Controlling the efficiency of excision repair. Mutat Res 485:3-13.
Hanawalt PC, Cooper PK, Smith CA. 1981. Repair replication schemes in bacteria and human cells. Prog Nucleic Acid Res Mol Biol 26:181-196.
Hanawalt PC, Cooper PK, Ganesan AK, Lloyd RS, Smith CA, Zolan ME. 1982. Repair responses to DNA damage: enzymatic pathways in E. coli and human cells. J Cell Biochem 18:271-283.
Howard-Flanders P, Boyce RP. 1966. DNA repair and genetic recombination: studies on mutants of Escherichia coli defective in these processes. Radiat Res Suppl 6:156-184.
Howard-Flanders P, Rupp WD. 1972. Recombination repair in UV-irradiated Escherichia coli, In: Molecular and Cellular Repair Processes (Beers Jr. RF, Herriott RM, Tilghman RC. ed), Johns Hopkins Medical Journal, Suppl. 1, p 212-225.
Howard-Flanders P, Rupp WD. 1981. Measurement of postreplication repair in prokaryotes, In: DNA Repair: A Laboratory Manual of Research Procedures, Vol. 1, Part B (Friedberg EC, Hanawalt PC. ed.) Marcel Dekker, New York p 459-470.
Howard-Flanders P, Rupp WD, Wilkins BM, Cole RS. 1968. DNA replication and recombination after UV irradiation. Cold Spring Harbor Symp Quant Biol 33:195-207.
Iyer VN, Rupp WD. 1971. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim Biophys Acta 228:117-126.
Jansz HS, Pouwels PH, Van Rotterdam C. 1963. Sensitivity to ultraviolet light of single- and double-stranded DNA. Biochim Biophys Acta 76:655-657.
Kato T. 1977. Effects of chloramphenicol and caffeine on postreplication repair in uvrAumuC and uvrArecF strains of Escherichia coli K-12. Mol Gen Genet 156:115-120.
Kato T, Shinoura Y. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet
14:121-131.
Kenyon CJ, Walker GC. 1981. Expression of the E. coli uvrA gene is inducible.
Nature 289:808-810.
Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage  . Microbiol Mol Biol Rev 63:751-813.
. Microbiol Mol Biol Rev 63:751-813.
Lin CG, Kovalsky O, Grossman L. 1997. DNA damage-dependent recruitment of nucleotide excision repair and transcription proteins to Escherichia coli inner membranes. Nucleic Acids Res 25:3151-3158.
Liu H, Cheng A, Wang TV. 1998. Involvement of recF, recO, and recR genes in UV-radiation mutagenesis of Escherichia coli. J Bacteriol 180:1766-1770.
Madiraju MV, Clark AJ. 1991. Effect of RecF protein on reactions catalyzed by RecA protein. Nucleic Acids Res 19:6295-6300.
Petit C, Sancar A. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 81:15-25.
Rupp WD, Howard-Flanders P. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31:291-304.
Rupp WD, Iyer VN, Zipser E. 1973. The reconstitution of chromosomal DNA in irradiated cells by post-replication recombinational repair, In: Advances in Radiation Research: Physics and Chemistry, Duplan JF, Chapiro A. ed. (Proceedings of the 4th Congress of Radiation Research, Evian-les-Bains, France, 1970) Gordon and Breach, New York p 39-50.
Rupp WD, Wilde III CE, Reno DL, Howard-Flanders P. 1971. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol 61:25-44.
Sargentini NJ, Smith KC. 1981. Much of spontaneous mutagenesis in Escherichia coli is due to error-prone DNA repair: Implications for spontaneous carcinogenesis. Carcinogenesis 2:863-872.
Sargentini NJ, Smith KC. 1989. Mutational spectrum analysis of umuC-independent and umuC-dependent gamma-radiation mutagenesis in Escherichia coli. Mutat Res 211:193-203.
Schendel PF, Fogliano M, Strausbaugh LD. 1982. Regulation of the Escherichia coli K-12 uvrB operon. J Bacteriol 150:676-685.
Sedgwick SG. 1975. Genetic and kinetic evidence for different types of postreplication repair in Escherichia coli B. J Bacteriol 123:154-161.
Sharma RC, Smith KC. 1985. A minor pathway of postreplication repair in Escherichia coli is independent of the recB, recC and recF genes. Mutat Res 146:169-176.
Sharma RC, Smith KC. 1987. Role of DNA polymerase l in postreplication repair: A reexamination with Escherichia coli 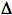 polA. J Bacteriol 169: 4559-4564.
polA. J Bacteriol 169: 4559-4564.
Shlaes DM, Anderson JA, Barbour SD. 1972. Excision repair properties of isogenic rec-mutants of Escherichia coli K-12. J Bacteriol 111:723-730.
Sigal N, Alberts B. 1972. Genetic recombination: The nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol 71:789-793.
Smith, KC. 2004. Recombinational DNA repair: the ignored repair systems. BioEssays 26:1322-1326.
Smith KC, Meun DHC. 1970. Repair of radiation-induced damage in Escherichia coli. I. Effect of rec mutations on post-replication repair of damage due to ultraviolet radiation. J Mol Biol 51:459-472.
Smith KC, Sharma RC. 1987. A model for the recA-dependent repair of excision gaps in UV-irradiated Escherichia coli. Mutat Res 183:1-9.
Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. 1999. UmuD'2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA 96:8919-8924.
Tseng YC, Hung JL, Wang TC. 1994. Involvement of RecF pathway recombination genes in postrepliction repair in UV-irradiated Escherichia coli cells. Mutat Res 315:1-9.
Wang TV, Smith KC. 1975. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol 156:1093-1098.
Wang TV, Smith KC. 1984. recF-Dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J Bacteriol 158:727-729.
Wang TV, Smith KC. 1985. Role of the umuC gene in postreplication repair in UV-irradiated Escherichia coli K-12 uvrB. Mutat Res 145:107-112.
Wang TV, Smith KC. 1986. Postreplicational formation and repair of DNA double-strand breaks in UV-irradiated Escherichia coli uvrB cells. Mutat Res
165:39-44.
Yarus M, Sinsheimer RL. 1964. The U.V.-resistance of double-stranded
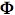 X174 DNA. J Mol Biol 8:614-615.
X174 DNA. J Mol Biol 8:614-615.
Youngs DA, van der Schueren E, Smith KC. 1974. Separate branches of the uvr gene-dependent excision repair process in ultraviolet-irradiated Escherichia coli K-12 cells; their dependence upon growth medium and the polA, recA, recB, and exrA genes. J Bacteriol 117:717-725.
Recent Articles
Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J; 2009 Apr 8;28(7):915-25.
Bordeianu G, Zugun-Eloae F, Rusu MG. The role of DNA repair by homologous recombination in oncogenesis. Rev Med Chir Soc Med Nat Iasi; 2011 Oct-Dec; 115(4):1189-94.
Fujinaka Y, Matsuoka K, Iimori M, Tuul M, Sakasai R, Yoshinaga K, Saeki H, Morita M, Kakeji Y, Gillespie DA,Yamamoto K, Takata M, Kitao H, Maehara Y. ATR-Chk1 signaling pathway and homologous recombinational repair protect cells from 5-fluorouracil cytotoxicity. DNA Repair (Amst); 2012 Mar 1;11(3):247-58
Ghodke I, Muniyappa K. Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae Mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins. J Biol Chem; 2013 Apr 19; 288(16):11273-86.
Helle F. Germ cell DNA-repair systems-possible tools in cancer research? Cancer Gene Ther; 2012 Apr; 19(4):299-302
Helmink BA, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu Rev Immunol; 2012 Apr 23; 30:175-202
Holkers M, de Vries AA, Gonçalves MA. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res; 2012 Mar; 40(5):1984-99
Izhar L, Ziv O, Cohen IS, Geacintov NE, Livneh Z. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proc Natl Acad Sci U S A; 2013 Apr 16;110(16):E1462-9.
Kumar A, Prameela TP, Suseelabhai R. A unique DNA repair and recombination gene (recN) sequence for identification and intraspecific molecular typing of bacterial wilt pathogen Ralstonia solanacearum and its comparative analysis with ribosomal DNA sequences. J Biosci; 2013 Jun; 38(2):267-78.
Lee B, Morano A, Porcellini A, Muller MT. GADD45α inhibition of DNMT1 dependent DNA methylation during homology directed DNA repair. Nucleic Acids Res; 2012 Mar 1; 40(6):2481-93.
Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res; 2008 Jan;18(1):99-113.
Liu J, Ehmsen KT, Heyer WD, Morrical SW. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit Rev Biochem Mol Biol; 2011 Jun;46(3):240-70.
Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol; 2005 Jul;57(1):97-110.
Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol; 2013;3:113.
Muñoz-Galván S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A. Distinct roles of mus81, yen1, slx1-slx4, and rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol; 2012 May; 32(9):1592-603.
Nickoloff JA, Brenneman MA. Analysis of recombinational repair of DNA double-strand breaks in mammalian cells with I-SceI nuclease. Methods Mol Biol; 2004;262:35-52.
Otón F, Martinez J, Mas-Torrent M, Garcia F, Alonso JC, Rovira C, Garcia R. Detection of the early stage of recombinational DNA repair by silicon nanowire transistors. Chiesa M, Cardenas PP, Nano Lett; 2012 Mar 14;12(3):1275-81.
Panico ER, Ede C, Schildmann M, Schürer KA, Kramer W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast; 2010 Jan;27(1):11-27.
Popławski T, Błasiak J. DNA homologous recombination repair in mammalian cells. Postepy Biochem; 2006;52(2):180-93.
Serrano MA, Li Z, Dangeti M, Musich PR, Patrick S, Roginskaya M, Cartwright B, Zou Y. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene; 2013 May 9; 32(19):2452-62.
Stankova K, Ivanova K, Mladenov E, Rosidi B, Sharma A, Boteva R, Iliakis G. Conformational transitions of proteins engaged in DNA double-strand break repair, analysed by tryptophan fluorescence emission and FRET. Biochem J; 2012 May 1; 443(3):701-9
Steger M, Murina O, Hühn D, Ferretti LP, Walser R, Hänggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak P, Gerrits B, Del Sal G, Zerbe O, Sartori AA. Prolyl Isomerase PIN1 Regulates DNA Double-Strand Break Repair by Counteracting DNA End Resection. Mol Cell; 2013 May 9; 50(3):333-43.
Thakur RS, Basavaraju S, Somyajit K, Jain A, Subramanya S, Muniyappa K, Nagaraju G. Evidence for the role of Mycobacterium tuberculosis RecG helicase in DNA repair and recombination. FEBS J; 2013 Apr; 280(8):1841-60.
Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, Burma S. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst); 2012 Apr 1; 11(4):441-8
Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A; 2013 May 7; 110(19):7720-5.
Walczak A, Rusin P, Dziki L, Zielinska-Blizniewska H, Olszewski J, Majsterek I. Evaluation of DNA double strand breaks repair efficiency in head and neck cancer. DNA Cell Biol; 2012 Mar; 31 (3):298-305
Wang G, Lo LF, Maier RJ. The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA Repair (Amst); 2011 Apr 3;10(4):373-9.
Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Danielsen JM, Yang YG, Qi Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell; 2012 Mar 30;149(1):101-12.
Weiner A, Zauberman N, Minsky A. Recombinational DNA repair in a cellular context: a search for the homology search. Nat Rev Microbiol; 2009 Oct;7(10):748-55.
Wibom C, Sjöström S, Henriksson R, Brännström T, Broholm H, Rydén P, Johansen C, Collatz-Laier H, Hepworth S, McKinney PA, Bethke L, Houlston RS, Andersson U, Melin BS. DNA-repair gene variants are associated with glioblastoma survival. Acta Oncol; 2012 Mar; 51(3):325-32
Williams AB, Hetrick KM, Foster PL. Interplay of DNA repair, homologous recombination, and DNA polymerases in resistance to the DNA damaging agent 4-nitroquinoline-1-oxide in Escherichia coli. DNA Repair (Amst); 2010 Oct 5; 9(10):1090-7.
Yeung PL, Denissova NG, Nasello C, Hakhverdyan Z, Chen JD, Brenneman MA. Promyelocytic leukemia nuclear bodies support a late step in DNA double-strand break repair by homologous recombination. J Cell Biochem; 2012 May; 113(5):1787-99
Zhong J, Liao J, Liu X, Wang P, Liu J, Hou W, Zhu B, Yao L, Wang J, Li J, Stark JM, Xie Y, Xu X. Protein phosphatase PP6 is required for homology-directed repair of DNA double-strand breaks. Cell Cycle; 2011 May 1; 10(9):1411-9.
10/11/08
06/02/11
03/25/13
07/20/14